Homopolymer Score
The Homopolymer score is applicable only for Roche/454 and Ion Torrent data. The Homopolymer score penalizes indels that are found in homopolymer regions because such indels are typically the result of sequencing errors. The penalty is higher for longer homopolymer regions because the likelihood of sequencing errors in such regions is also higher. The software first determines which length of homopolymer region is present more often (A) and which length is present less often (B). If A or B is < 3, then the value for the Homopolymer score set to one; otherwise, the score is calculated according to the following:
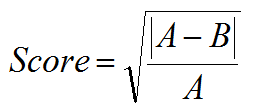
For example, deletion from four bases to three bases that occurs less than half of the time, where A = 4 and B = 3 results in a score of 0.5, which reduces the Overall Mutation score.