To split files
You use the Split files option to split a single .fasta or .fastq file into multiple .fasta or .fastq files. This is a useful option if a single sample file is taking considerable memory to analyze and you would like to carry out a series of smaller and faster analyses.
1. In the Input pane, click Add to browse to and select the .fasta file or .fastq file that is to be split into multiple files.
2. In the Settings field, enter the maximum acceptable size for each partition in MB.
3. In the Output field, you can leave the default value for the location of the output files as is (the default value is the directory path for the input file), or you can click Set to select a different location.
4. Optionally, before you process the files, click Save to save the settings that you have specified to a Settings file (.ini file).
You can always load this file at a later date and process other data files according to the saved settings in the file. |
5. Click OK.
A message opens when the process is completed.
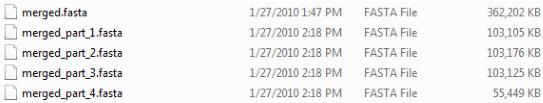
The single file is split into “x” number of equally sized partitions, with any remainder contained in a smaller file. For example, for a 5.5 KB file with a partition size of 1 KB, six files are produced—five 1 KB files and one 0.5 KB file. As shown in the figure below, the name for each partition is based on the name of the split file and is appended with the phrase “_part.” In addition, the partitions are numbered sequentially.
Multiple .fasta files created by splitting a single .fasta file