To build a BED file with the BED File Builder tool
1. On the Geneticist Assistant main menu, click Tools > Build BED file.
The BED File Builder dialog box opens.
2. Do one of the following:
• To manually specify the gene name, in the Gene Name field, enter the name for the gene that is in the panel, and then click Add Gene. Repeat this step for each gene that is in the panel.
As you enter the gene name, a list of all genes in the database that meet the search criteria is dynamically updated. You can select the gene from this list. The search is not case-sensitive, but the search string must begin with the gene name. For example, a search string of “as” returns ASH1L-AS1 but a search string of “as1” does not. |
• To load the gene names from a file, click the Browse button  to open the Select a gene file dialog box, browse to and select the correct file, and then click Load Genes.
to open the Select a gene file dialog box, browse to and select the correct file, and then click Load Genes.
 to open the Select a gene file dialog box, browse to and select the correct file, and then click Load Genes.
to open the Select a gene file dialog box, browse to and select the correct file, and then click Load Genes.The file must be a text file with one line per gene. Optionally, you can specify a transcript for each gene, with a tab between the gene name and the transcript. |
The following information is displayed for all the different transcripts that are available for the gene in the top pane of the dialog box—Region Name, Chromosome, Region Start position, Region End position, and Region Length.
3. If you are building a BED file for genes that are associated with a specific phenotype, then do the following; otherwise, go to Step 4.
In the Phenotype field, enter a search string for the phenotype. As you enter the search string, a dropdown list opens. The list is dynamically updated with the phenotypes that are found in the Human Phenotype Ontology database that match the search string.
The search string is not case-sensitive, and the string can be found anywhere in the search results. For example, a search string of “thyroid” returns phenotypes of thyroid adenoma and secondary hyperthyroidoism. |
4. Select the mode, which determines the information that is available for creating the BED file.
Mode | Description |
|---|---|
Exon | The default mode. You can include only the coding regions in the BED file, only the untranslated regions, or both. Introns are not included in Exon mode. |
Transcript mode | Adds the full transcript, including introns, to the BED file. |
All Exon Mode | Includes all the specified exons from all the selected transcripts in the BED file. You can include only the coding regions in the BED file, only the untranslated regions, or both. Introns are not included in Exon mode. |
5. Do one of the following:
• For Exon mode and All Exon mode—Select a transcript, or click the Expand icon 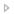 next to the transcript, and then select the coding region and/or untranslated regions for building the BED file.
next to the transcript, and then select the coding region and/or untranslated regions for building the BED file.
 next to the transcript, and then select the coding region and/or untranslated regions for building the BED file.
next to the transcript, and then select the coding region and/or untranslated regions for building the BED file.BED File Builder tool, Exon mode
• For Transcript mode—Select a transcript.
6. If you are building a BED file for genes that are associated with a specify phenotype, then click Add Gene; otherwise, go to Step 7.
The appropriate regions are added for the BED file based on the selected Mode. See Step 4.
7. Optionally, do any or all of the following:
Option | Steps |
|---|---|
To open the Preview pane and view a preview of the BED file | Click Preview. The information for each region (Chromosome, Region Start, Region End, Region Name, Score, and Strand) is displayed in the preview. Note: For detailed information about the fields that are displayed in the preview, see https://genome.ucsc.edu/FAQ/FAQformat.html#format1. You can leave the Preview pane open, or you can close it after viewing the BED file. If you leave the pane open, and then make any changes to any of the BED file settings, you must click Refresh to update the preview. |
To adjust the region start and end positions | Enter a different Region Start and/or Region End value (the default value is zero). For example, to include 20 bases at the end of each exon, enter 20 for both the Region Start value and the Region End value |
To apply the current settings (the selected mode and the Start/End Region values) to all genes, including any previously added genes | 1. Click Apply Settings to All. An Overwrite warning message opens, indicating that all your gene-specific settings will be lost and asking you if you want to continue. 2. Click Yes to close the message and overwrite all the current gene-specific settings with the current gene settings. |
To immediately use the BED file as a panel | 1. Click Import as Panel. The Panel Name dialog box opens. 2. Enter the name for the panel, and then click OK. The Panel Name dialog box closes, and the Manage Panels dialog box opens. The full directory path to the imported panel, including the panel name, is displayed in the Import from Panel File field. 3. Continue to Importing a Panel. |
8. Click OK.
The Save As dialog box opens. By default, the BED file is named default.bed.
9. Edit the name of the BED file as needed, and save it in a location of your choosing.