Forensic & Medical Mitochondrial DNA Analysis
Developed in collaboration with leading forensic and medical laboratories, GeneMarkerHTS software provides rapid analysis of multiple samples using consensus and motif alignments:
- Unique motif alignment technology
- Automates the requirements set by The DNA Commission of the International Society for Forensic Genetics
- Fulfills the forensic alignment recommendations.
- Provides recognition and proper assignment of motifs and indels consistent with phylogenetic and forensic considerations
- Consensus alignment
- To the preloaded Revised Cambridge Reference Sequence or to other references
Motif alignment reduces manual edits for forensic alignment
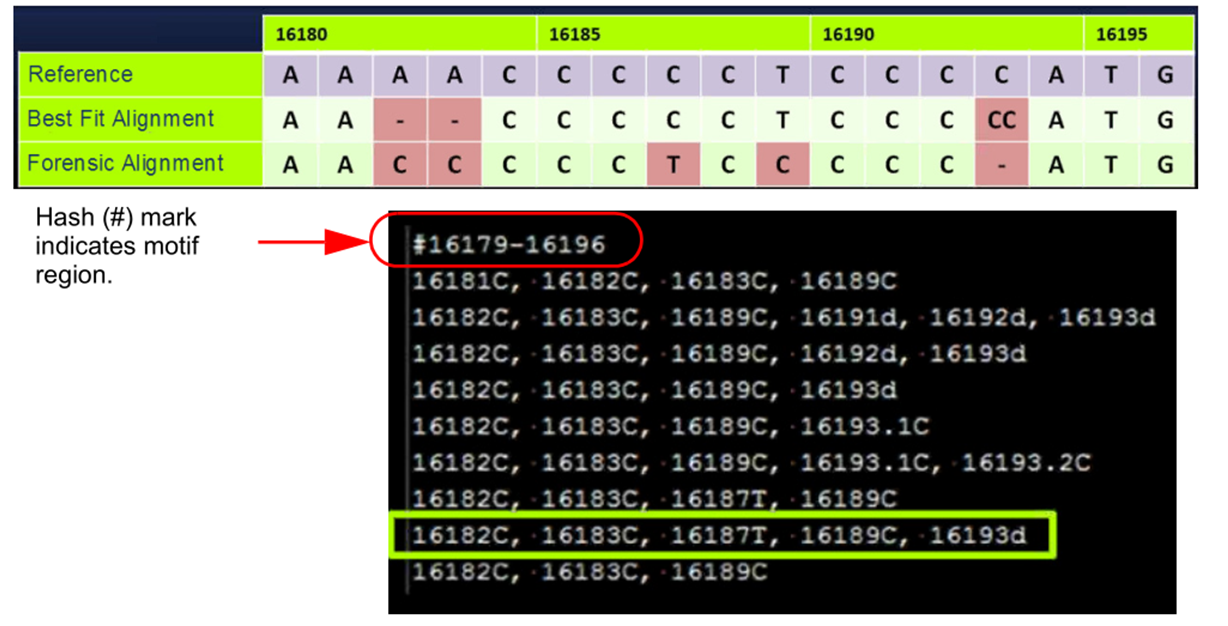
Figure 1: GeneMarkerHTS software has an extensive, preloaded forensic motif file as well as a motif editor to assist labs in adding new motifs.
Automatic Features Include:
- Three alignment algorithms that best suit the analysis needs:
- Initial Alignment to a reference genome (Revised Cambridge Reference Sequence is preloaded into GeneMarkerHTS)
- Adjust alignment with motifs to ensure the alignment is consistent with forensic variant reporting
- Adjust alignment using the consensus; improving alignment around indels
- Analysis across the origin
- Forensic or standard nomenclature
- Maintain health privacy with an administrative option that blocks display of disease associated positions
Analysis Results Include:
- Consensus sequence, Variants, SNPs, Indels
- Depth of coverage graphics
- Consensus sequence aligned to reference (IUPAC nomenclature)
- Whole mtDNA genome, spanning the origin
- Specified areas of interest, such as control region, HV1, HV2
- Read pile-up (with depth and direction indicators)
- Compare multiple samples in single view
- Synchronized view - scroll and zoom multiple samples
- Comparison viewer - table with sample-to-sample and variant composition
GeneMarkerHTS Software Viewing Options:
- Users can customize color-coding for nucleotides and highlight edits
- ‘Global’ and ‘Zoom’ views for the whole mtDNA genome alignment
- The Global View shows the depth of coverage with forward read coverage in blue and reverse read coverage in red
- The Zoom View allows users to zoom-in on the range displayed in the pile-up
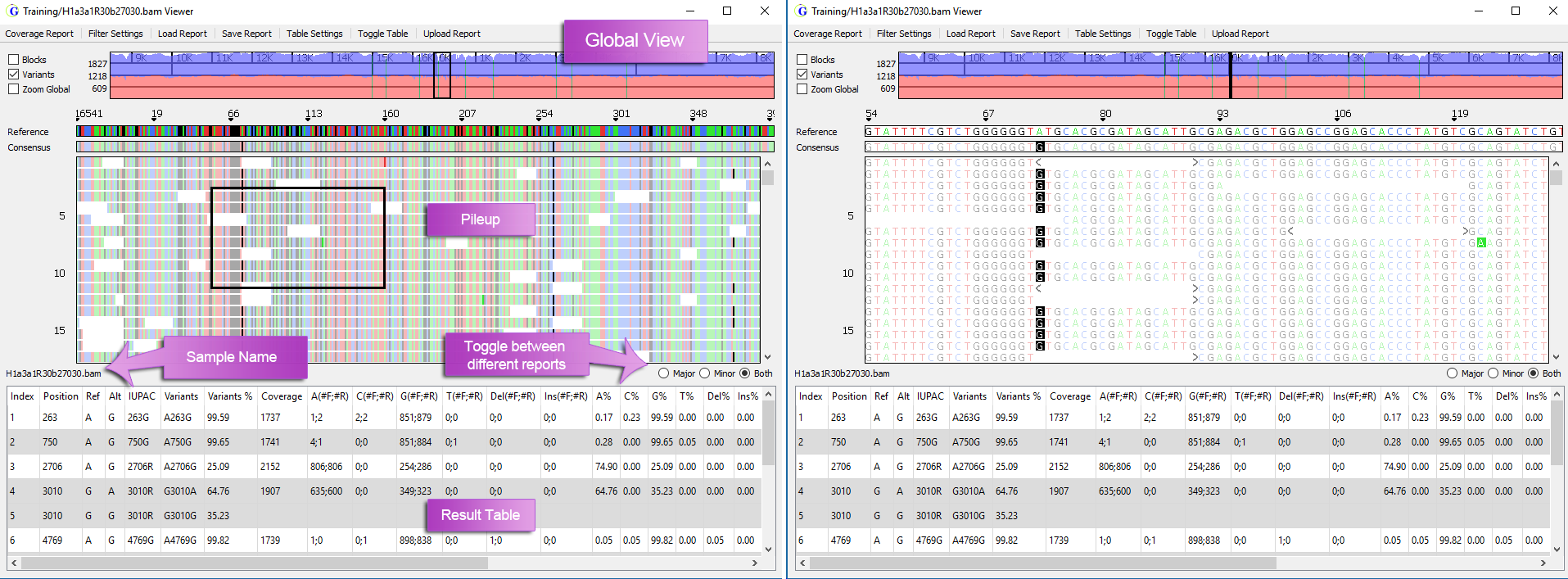
Figure 2: Global view (left) and Zoom view (right).
View up to 4 samples simultaneously
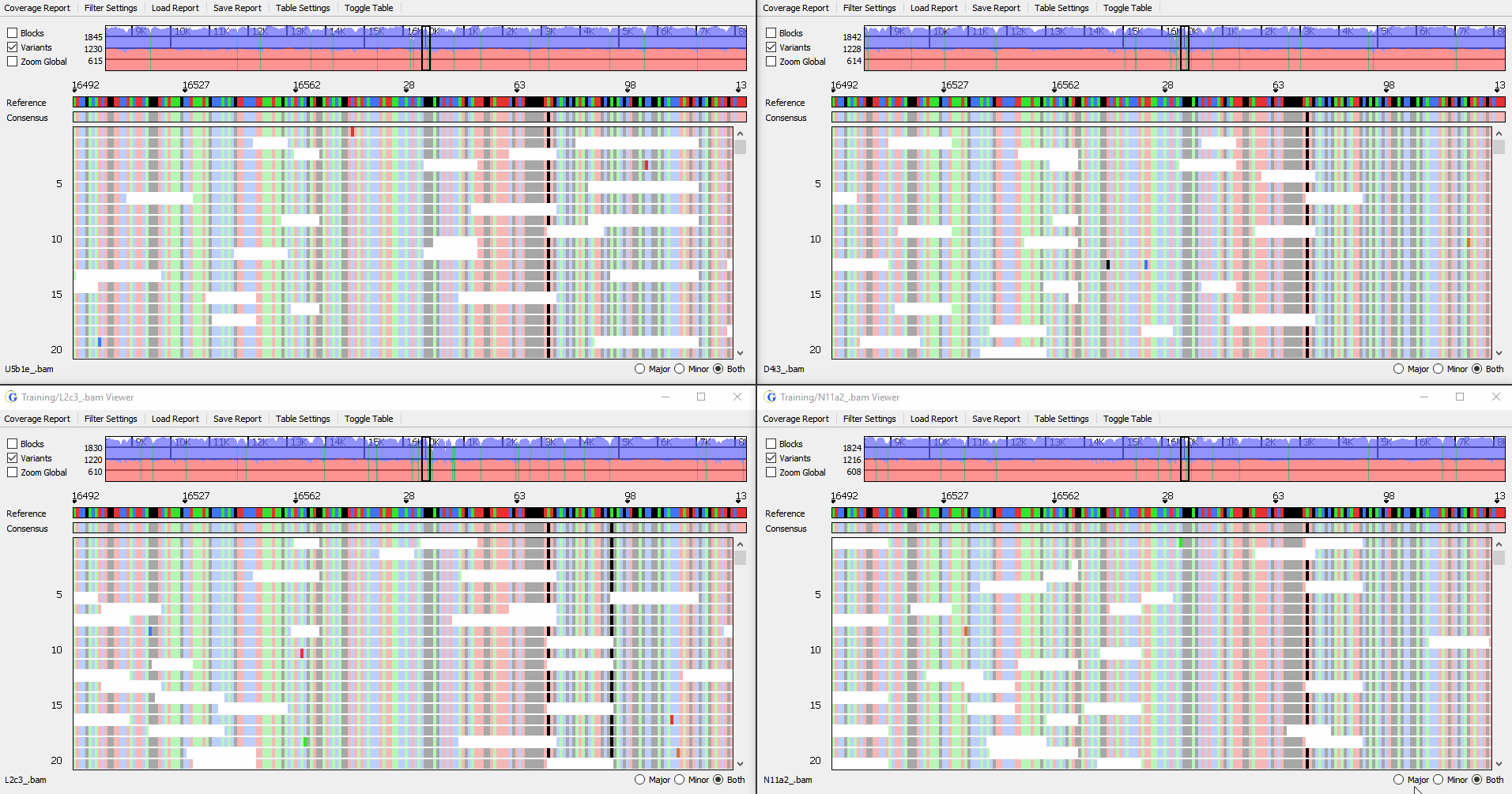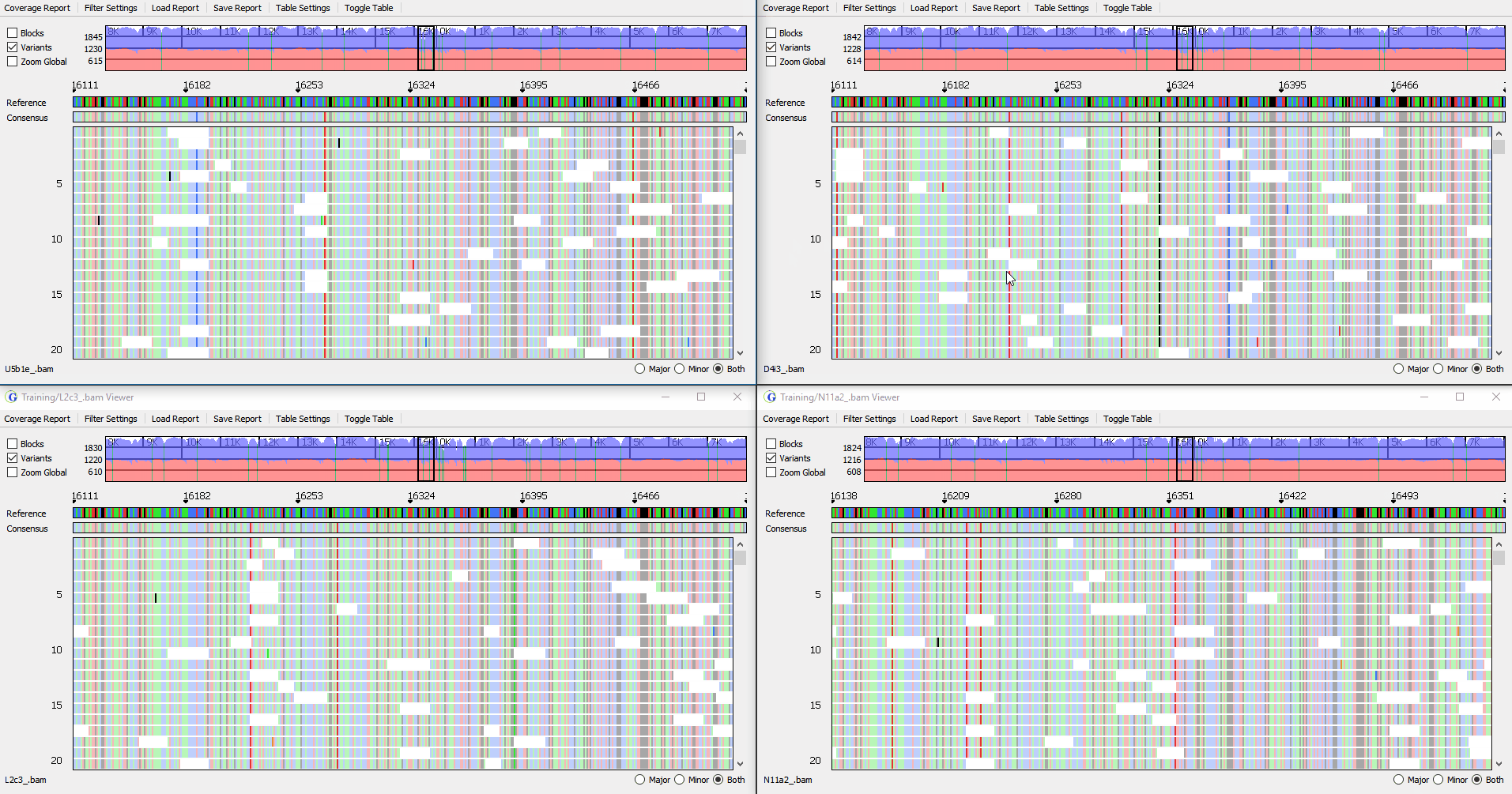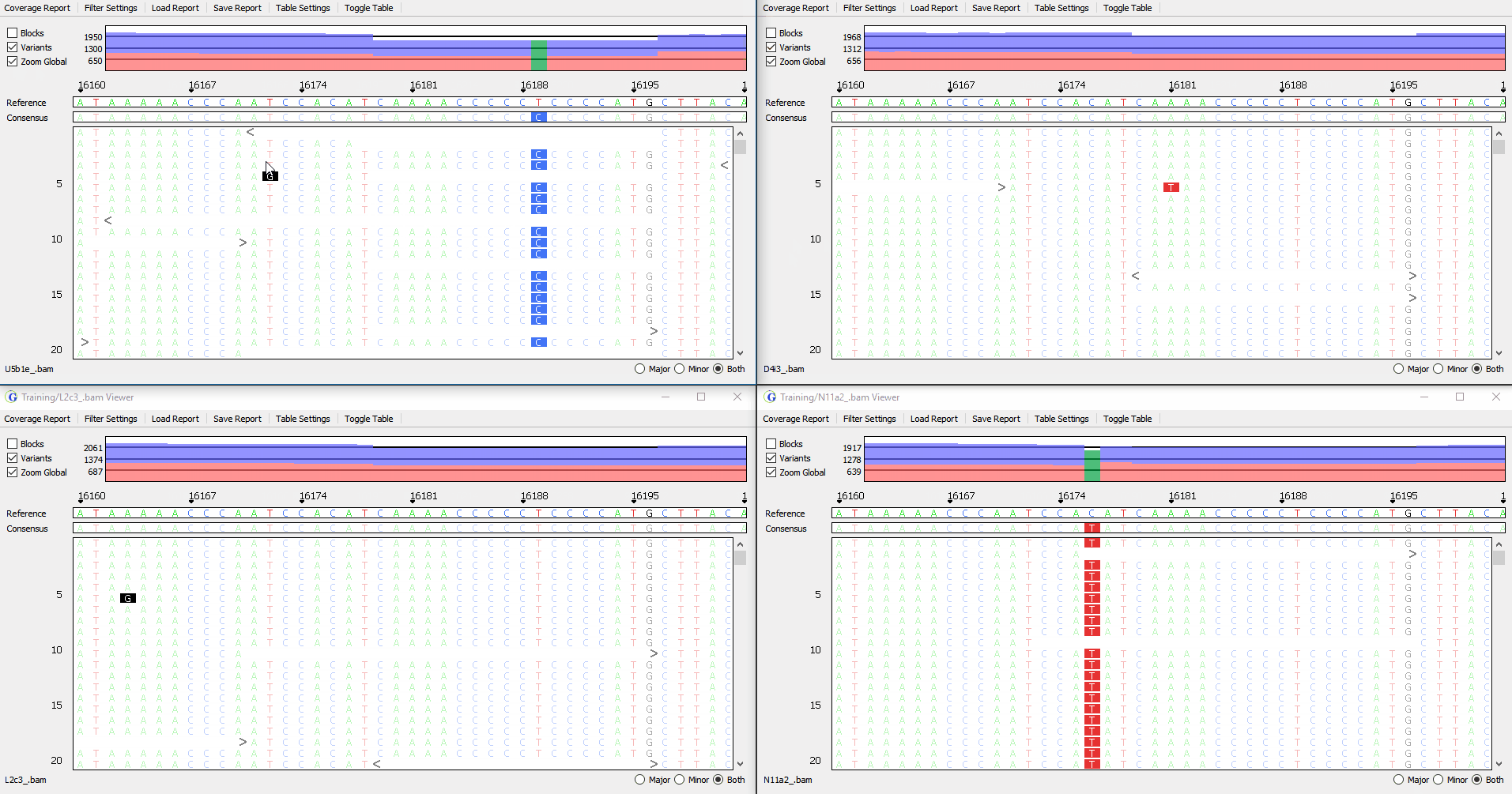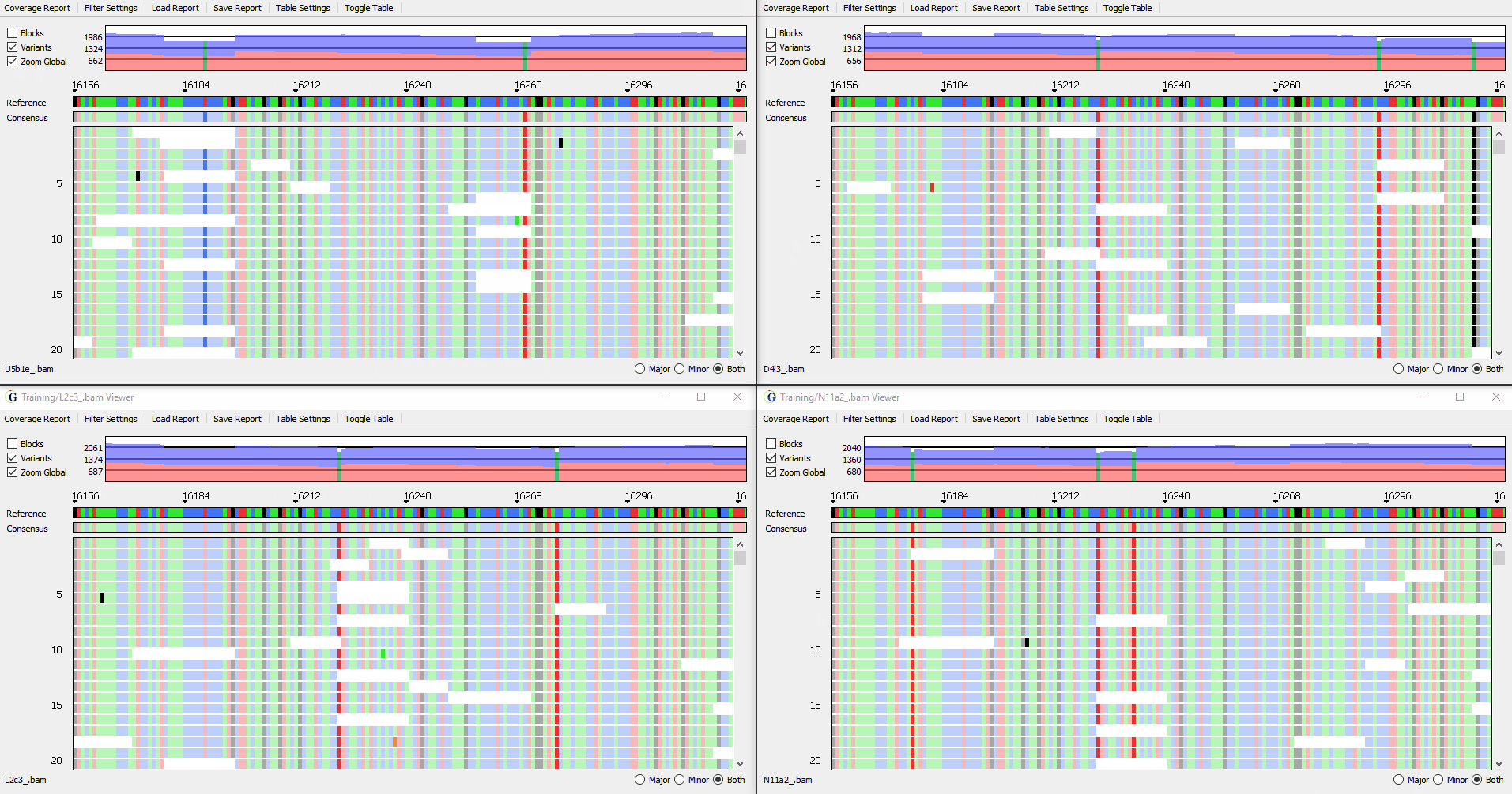
Figure 3: Synchronized viewing of multiple samples
Compare analysis results of multiple samples
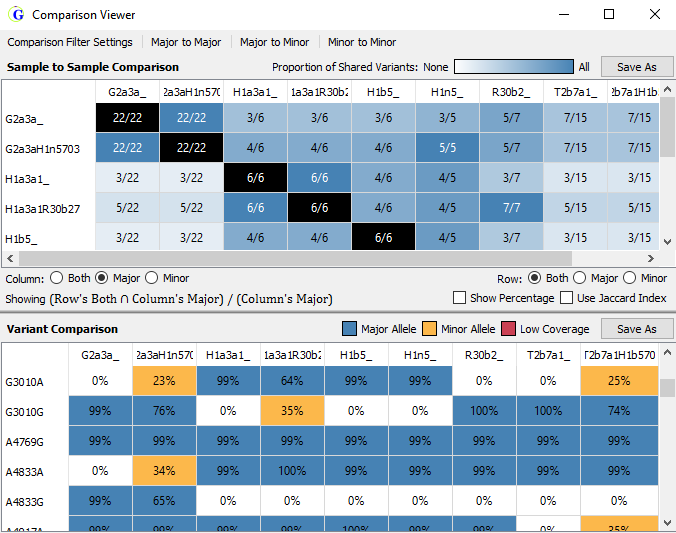
Figure 4: Sample-to-sample comparison (top) and variant comparison of all samples in the project (bottom)
GMHTS Software can Export a Variety of Reports:
- Consensus sequence
- Variant reports – SNPs, heteroplasmy, and insertions and deletions
- Variant reports are EMPOP (EDNAP mtDNA Population Database) compatible
Minimum Recommended processing hardware:
- 64-bit Windows OS 12 GB RAM 2.4GHzDual Quad Core Processor.
Reference Material:













