Whole Genome Mitochondrial DNA Analysis
NextGENeLR software can be used for the analysis of whole genome mitochondrial DNA sequencing using long read data such as data from Pacific BioSciences systems. Pacific Biosciences SMRT (single molecule, real-time) sequencing, produces reads of dozens of kilobases in length, enabling the sequencing of up to the entire mitochondrial genome in one pass.
Whole genome mitochondrial DNA sequencing can be analyzed using NextGENeLR software for:
- Detection of structural variants
- Haplotyping, including mixed samples
- Detection of SNVs and indels, including low frequency variants
In conjunction with long read lengths of dozens of kilobases, NextGENeLR provides a powerful solution for the detection of structural variants. NextGENeLR software allows the accurate haplotyping of long read data by implementing accurate alignment and comparing aligned reads to reference haplotypes. Haplotypes can also be discerned from mixture samples, with detection as low as 0.1% frequency. NextGENeLR software can detect SNVs and small indels at frequencies as low as 2%.
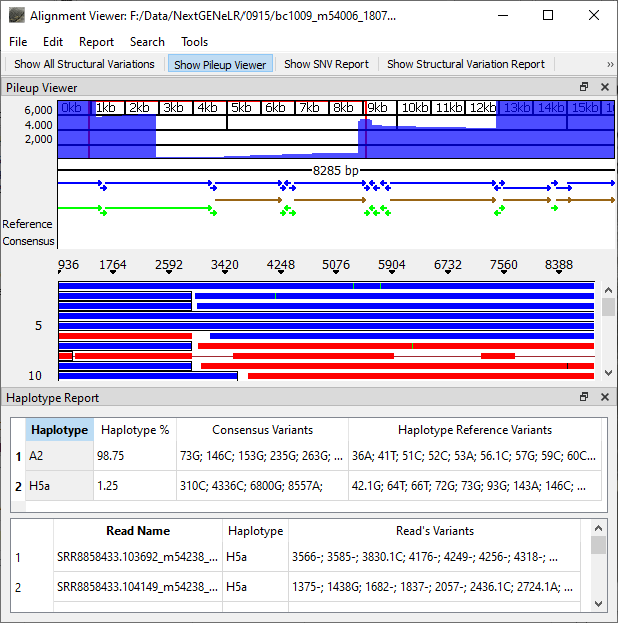
Figure 1: NextGENeLR Alignment Viewer with Haplotype Report displayed. The Haplotype Report for this sample indicates a mixture is present with the minor haplotype present at 1.25% frequency. The bottom section of the report lists the haplotype and variants present for each individual read in the sample.
Application Notes:
Trademarks property of their respective owner













