CASE SPECIFIC PANEL CONSTRUCTION TOOL FOR AUTOMATED IDENTIFICATION OF INFORMATIVE ALLELES IN POST-TRANSPLANT ANALYSES
Applying panels to sized data is required for consistent allele calls and accurate calculation of peak ratios. Panels are XML files that contain information on all fragments that may result from the PCR reactions. ChimeRMarker® software uses two types of panels: Genotyping Panels and Chimertyping Panels.
Genotyping panels determine the allele calls of recipient and donor. Panels for commercial Human Identity kits are pre-loaded in ChimeRMarker software, or if custom primer sets are used, the Panel Editor allows rapid creation of genotyping panels.
Case-specific Chimertyping Panels are based on the allele calls of the recipient and donor(s) and are made automatically in the Panel Editor. Once made they can be applied to all later samples for that case, providing accuracy and time savings to longitudinal monitoring for engraftment. The Chimertyping panel enables ChimeRMarker software to automatically designate donor, recipient and shared peaks in post BMT samples.
Panel files (including Identifiler®, GlobalFiler™ 6-Dye™, PowerPlex®16, PowerPlex®ESI, GenePrint® 24, PowerPlex®Fusion 5 and 6 Dye, Investigator®24Plex) and size standards (including GeneScan™500-GS600 and Internal Lane Standard ILS500-ILS600) are pre-loaded in ChimeRMarker software. Filtering parameters of each locus can be customized in the panel editor, including: Minimum Heterozygous and Homozygous RFUs for Analytical Range and Stochastic Ranges, and percentages for Heterozygous Imbalance and Stutter. Size call quality is confirmed with linear plots for each file and quality statistics for disabled fragments, as is recommended for GeneScan™500LIZ®, and options to save or print the Sizing Quality Report. ChimeRMarker software imports files from all ABI®PRISM, SeqStudio, and Promega Spectrum Compact CE Systems.
Genotyping Panel
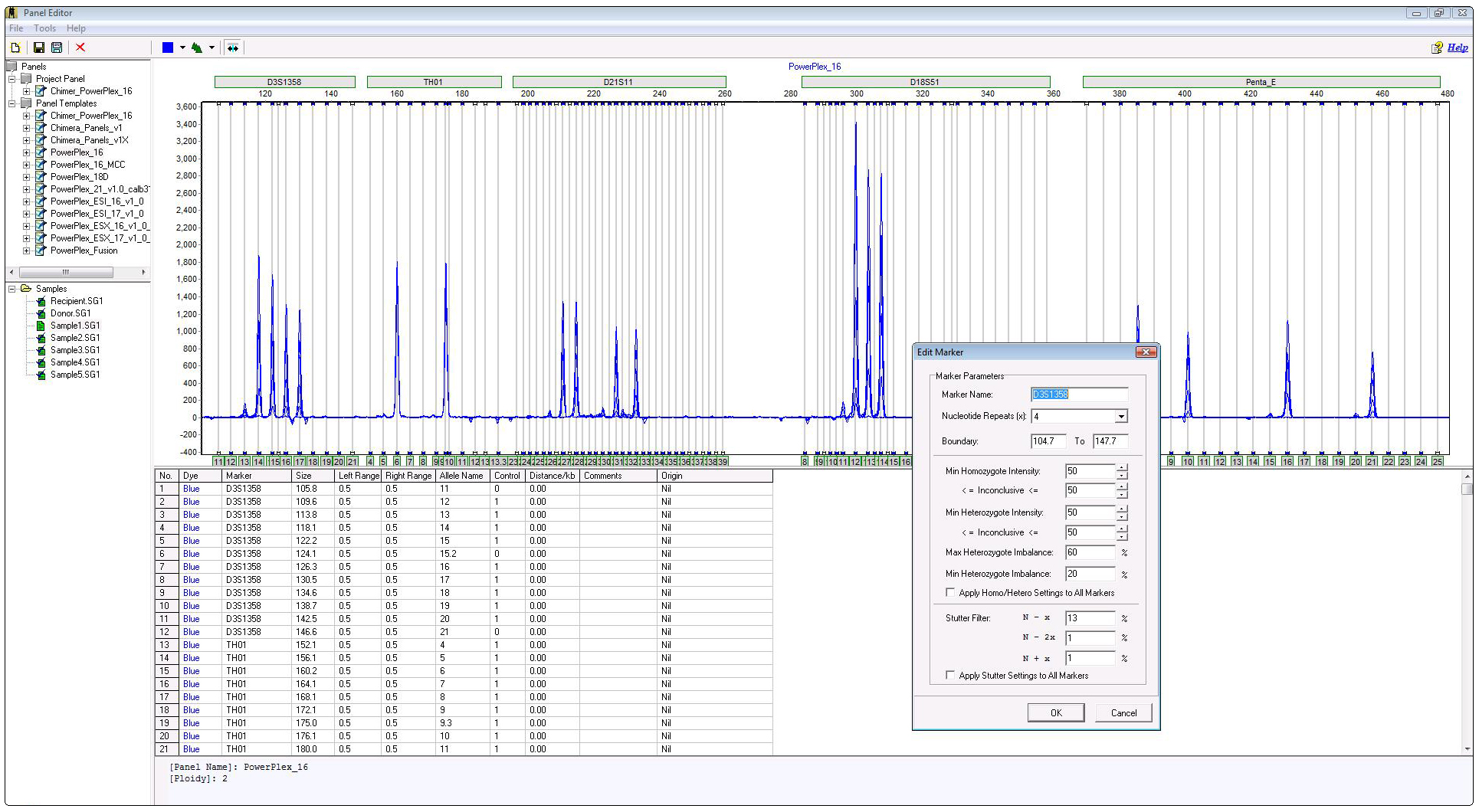
Figure 1: Genotyping panel contains all fragments that may result from the PCR and contains several quality flagging parameters that increase accuracy of allele calls.
Chimertyping Panel
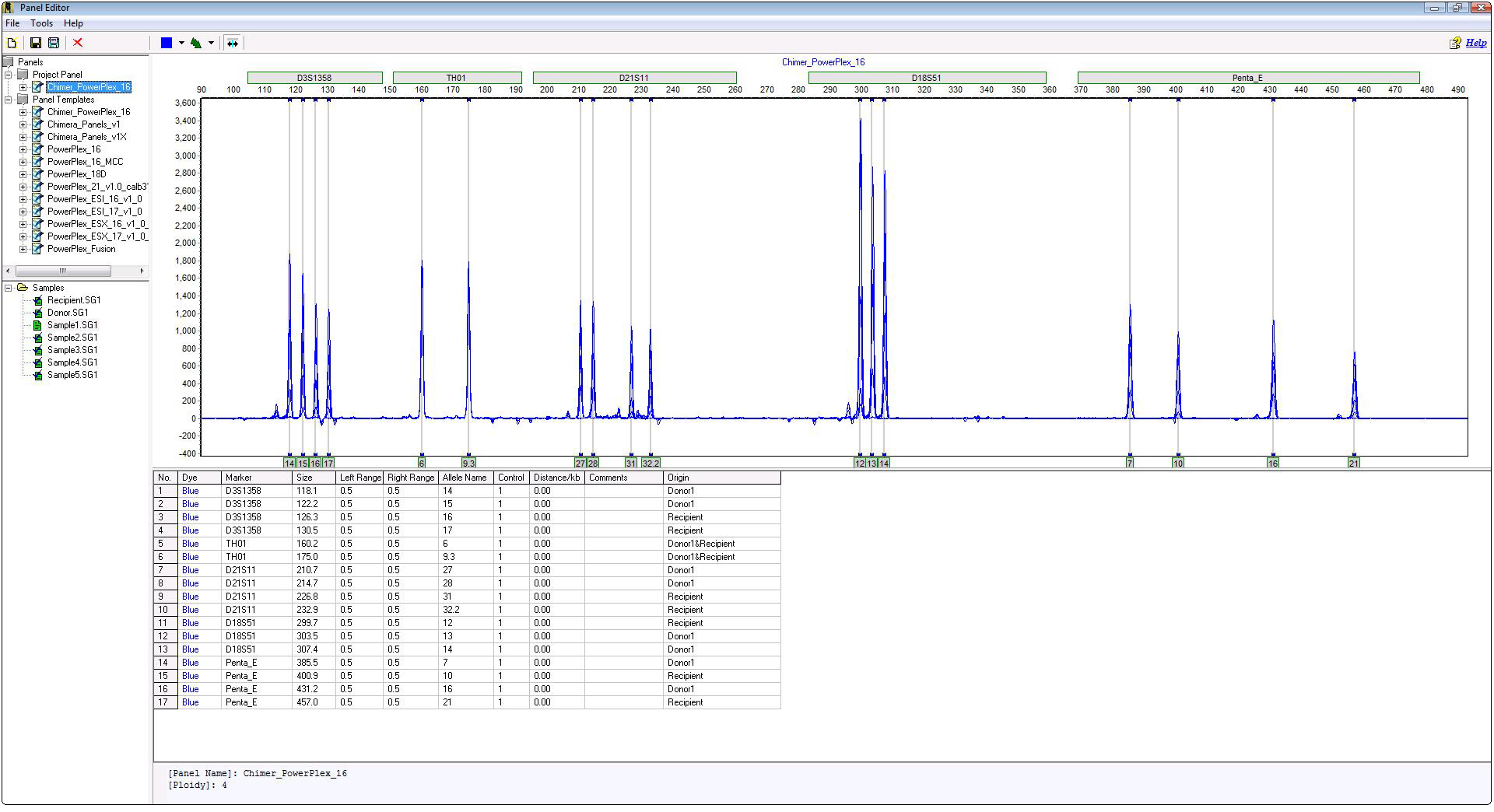
Figure 2: Chimertyping panel, which is case specific, identifies the origin (donor, recipient or shared) of each peak.













