Geneticist Assistant® NGS Interpretative Workbench
Variant Interpretation * Variant Tracking * Coverage Confirmation Save Time & Resources * Compatible with data from all NGS systems * Targeted Panels and Whole Exome Sequencing
Developed in collaboration with the Laboratory Medicine, Information Technology, and Health Science Research departments of Mayo Clinic, Geneticist Assistant NGS Interpretative Workbench is a unique tool for the management, control, visualization, functional interpretation, and knowledge database of next-generation sequencing whole exome data for the purpose of identifying pathogenic variants. Geneticist Assistant may also be used to target specific gene panels.
Geneticist Assistant is compatible with data processed from all leading next generation sequencing platforms such as those from Ion Torrent and Illumina. The program accepts standardized BAM and VCF files, includes prediction information from SIFT, PolyPhen2, LRT and Mutation Taster as well as conservation scores data from PhyloP, GERP++, SiPhy and population frequencies from ExAC, GnomAD, and Exome Variant Server. Additionally, information from proprietary custom databases and from the COSMIC database of somatic mutations are easily imported into the workbench.
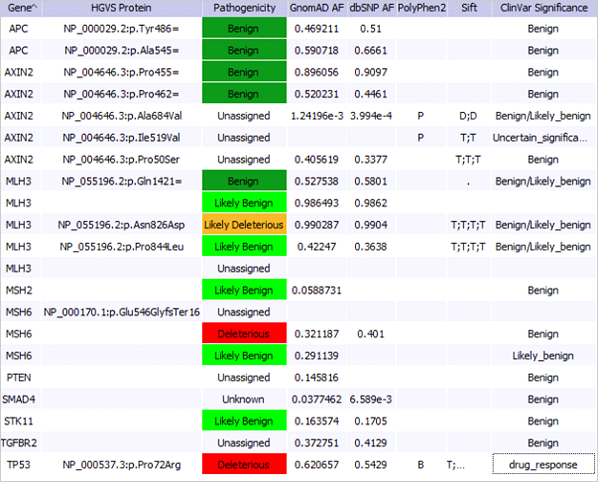
Sample’s Variants: Example configuration of variant table. Columns can be customized to show the fields needed. Variant pathogenicity is set by the user and shown in other samples.
Geneticist Assistant software includes valuable tools for variant filtering, sample comparisons - including trio and family comparisons - custom PDF report design, and review of customizable quality metrics.
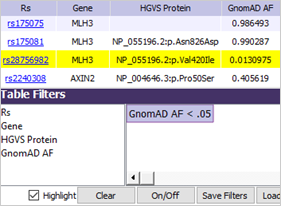
Filtering: The sample’s variants have been set to highlight only variants with GnomAD allele frequency of less than 0.05. Deselecting the Highlight option would display only those variants that match the filter.
Geneticist Assistant can also be integrated with NextGENe to provide a complete, seamless pipeline from alignment and variant calling to variant interpretation.
Compatible Databases
Functional Prediction Information:
SIFT, PolyPhen2, LRT, Mutation Taster, FATHMM, CADD & Mutation Assessor
Disease Association:
ClinVar, OMIM & COSMIC
Conservation Scores:
PhyloP, GERP++, phastCons & SiPhy
Population Frequencies:
1000 Genomes, GnomAD, ExAC, and Exome Variant Server
Additionally, information from proprietary databases such as Alamut and LOVD is easily accessible through embedded links. Information from other publicly available databases is also easily imported into the workbench.
Unique tools, including Quality Metrics Assistant, in conjunction with NextGENe software can form a completely automated informatics pipeline.
Webinars:













