Sample Comparison
Geneticist Assistant® Interpretive Workbench can be used to create comparisons between samples, including comparisons of family members based on relationship and phenotype information, as well as comparisons of samples without family relationships.
The “Family Comparison” function is a useful tool for identifying variants that are consistent with a defined inheritance pattern when samples for related individuals have been sequenced in order to obtain a refined list of variants of interest. Comparison results are displayed in a Family Comparison tab which displays only variants that match the inheritance pattern selected.

Family Comparison: Family comparison results show the allele frequency and coverage values for each sample included in the comparison along with pedigree symbols defining each sample.
The Family Comparison tab displays the results of the family based comparison, showing sample specific values for allele frequency, and optionally coverage and read balance for each sample on the left side. Samples are identified using pedigree symbols and sample name, relationship and phenotype can be shown by hovering the mouse over any of the sample columns.
To utilize this function VCF, and optionally BAM files, can be imported for multiple family members. Geneticist Assistant’s patient import function can be used to import patient IDs, phenotypes and relationship information for all related patients. Imported samples can then be automatically associated with the appropriate patient ID. The Family Comparison can be used when at least two parents and one child are available. Any number of children can be included in the comparison.

Family Comparison Settings: Options for the Family Comparison function, including relationships and phenotype, are automatically selected based on patient information in the database, when provided. Options can also be manually defined using the dropdown menus.
Specialized comparison options are also available when comparing samples without family relationships. The comparison can be created to look for shared variants in a specified number of samples, variants with allele frequency differences across the samples, within a specified range, or variants where the allele frequency in the samples is within a certain range.
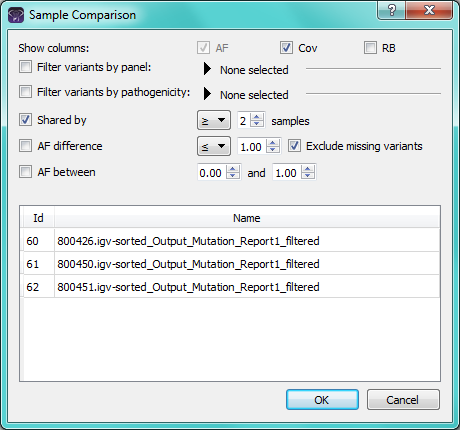
Sample Comparison Settings: Options are available to report variants shared by a set number of samples or with specified allele frequency requirements, as well as to optionally filter by panel or pathogenicity.













