MATERNAL CELL CONTAMINATION (MCC) DETECTION AND QUANTIFICATION
Cahorionic villus (CV) and amniotic fluid (AF) are common cell sources for prenatal molecular analysis of genetic disorders, such as aneuploidy/trisomy or cystic fibrosis. These samples may be contaminated with maternal tissue or maternal hematopoietic cells, decreasing the accuracy of genetic testing. The analyst must be confident that the sample used for analysis does not contain any maternal cell contamination. MCC analysis also serves as an internal quality assurance measure to ensure that the biological mother is matched with her concordant fetus(es), minimizing the possibility of sample mix-up. MCC analysis is a key requirement for umbilical cord blood banking amid concerns that maternal T cells may elicit GvHD post-UCB transplant. ChimeRMarker® software has a user-friendly interface, provides accurate genotyping of maternal and fetal samples and uses rigorous mixture analysis algorithms to automatically detect and quantify MCC. ChimerMarker software is compatible with data from all major capillary electrophoresis systems, including but not limited to ABI®PRISM, Applied Biosystems® SeqStudio, Hiachi High-Tech Compact CE Sequencer DS3000, Promega Spectrum Compact CE Systems and Syntol Nanophore® 05 genetic analyzers, and custom primers or commercially available human identification chemistries for STR genotyping (including, but not limited to Identifiler®, GlobalFiler™ 6-Dye™, PowerPlex®16, PowerPlex®ESI, GenePrint® 24, PowerPlex®Fusion 5 and 6 Dye, Investigator®24Plex).
Contact info@softgenetics.com to request a free trial and online training.
Maternal Cell Contamination (MCC) Application in ChimeRMarker software Provides:
- Accurate Size and Allele Calls
- Automated Detection and Quantification of MCC
- Time Efficiency …. Analyze Multiple Cases in One Project
- Create individual Case Reports - Quantification Follows MCC Best Practice Guideline (USA and European)
- Reduce Errors …. No data transfer - Allele Calls and Linked MCC Quantification Application
Dialog Boxes to Easily Set MCC Analysis Parameters
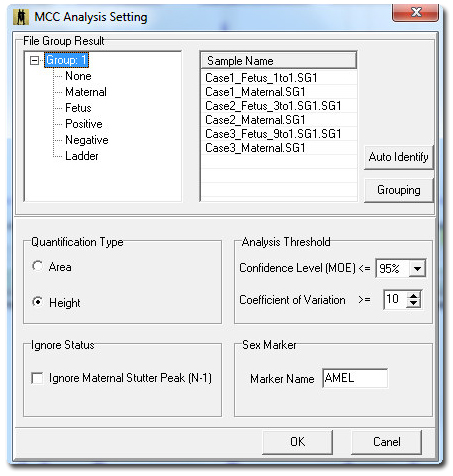
Figure 1: Maternal Cell Contamination analysis dialog window contains parameters to quantify in area or height, and to ignore stutter peak, gender marker, statistics and grouping options. Select Grouping to activate the File Group Editor.
The File Group Editor is used to group individual fetal samples to their respective maternal samples. After grouping of each case, the auto identify button can further classify each maternal and fetus sample into their respective category within each group.MCC Results
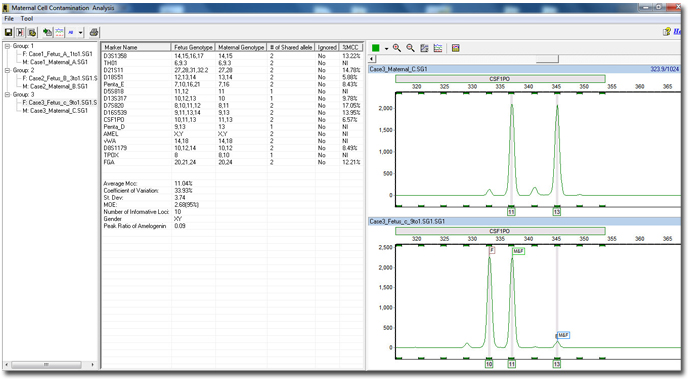
Figure 2: By grouping and auto identifying each sample, the software can analyze each fetus sample by comparing the alleles to their respective maternal sample. Each individual patient or group is defined in the browser pane on the left. The middle pane shows all calculated results, genotypes and parameters. The electropherograms identify each allele in the fetus sample as either a fetus, maternal, or shared allele for easy reviewing.
Summary Report for Each Case with Custom Header and Authorization Box
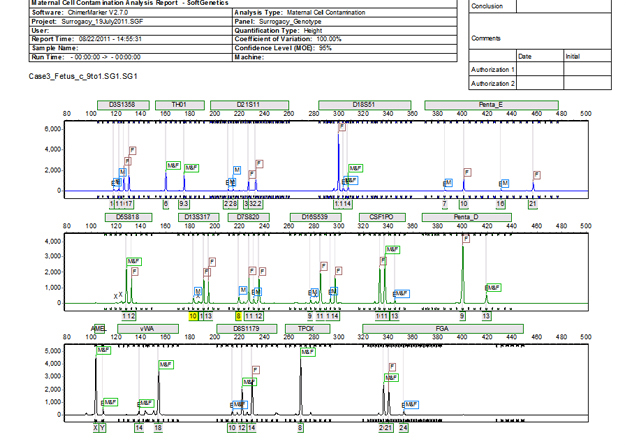
Figure 3: ChimeRMarker software's final MCC report consists of a two-page summary. The first page contains the electropherograms with all alleles identified along with a customer header that displays both the analysis information, institution information, and sample information.
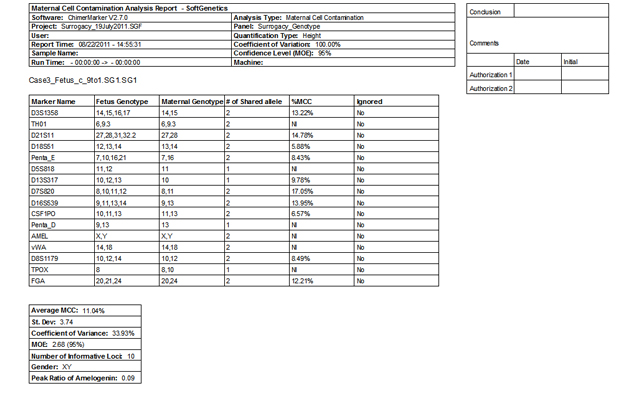
Figure 4: The second page of ChimeRMarker software's final MCC report consists of a two-page summary contains the quantification, statistics, and genotypes detected in the fetus and maternal sample.
Webinars:













