Time Saving, Quality Evaluation tools in GeneMarkerHID Software include an Embedded Contamination Check
The European Network of Forensic Science Institutes (ENFSI), the FBI Laboratory’s Scientific Working Group on DNA Analysis Methods (SWGDAM), and the Australian Biology Specialist Advisory Group (BSAG) issued a joint statement: “Because the sensitive PCR technique replicates any and all of the DNA contained in an evidence sample, greater attention to contamination issues is necessary.” The groups advocate for all forensic DNA laboratories to maintain an elimination database for screening DNA results. The results of a 2015 study by Séguin et al. published in Forensic Science International: Genetics, illustrates the importance of including genotypes of all crime scene workers (police, laboratory and field employees) in an elimination database. Typically, labs will compare profiles in spread sheets with macros to detect profile-to-profile and profile-to-staff member contamination, which is time consuming and error prone.
GeneMarkerHID software enables the analyst to easily and automatically perform profile comparisons to check for contamination, eliminating data transfer to a separate database or spreadsheet/macro. GeneMarkerHID software is compatible with major CE and Rapid Instrument data files, Windows® Operating Systems that are within Microsoft Extended Support and all major commercial human identity chemistries.
Cross contamination of a DNA sample can occur from several sources. Sources of potential staff member-to-case profile contamination include touch DNA (property crime), cold cases, and handling by multiple individuals in the chain of custody process. Examples of profile–to–profile contamination within a project include: low template DNA from surface swabs, overloading or spectral overlap issues from capillary electrophoresis and sample mishandling. GeneMarkerHID software has built-in contamination detection.
The contamination check provides an immediate screening for contamination:
- Profile-to-Profile contamination within a project
- Staff member genotype inclusion in profile(s)
View the ISHI 2016 poster: Identify QC Problems Automatically
Figures 1 and 2 provide a summary of the Contamination Check in GeneMarkerHID software and are followed with details on the equation and features of the contamination check.
Contamination Check Setting Options and Result Tables
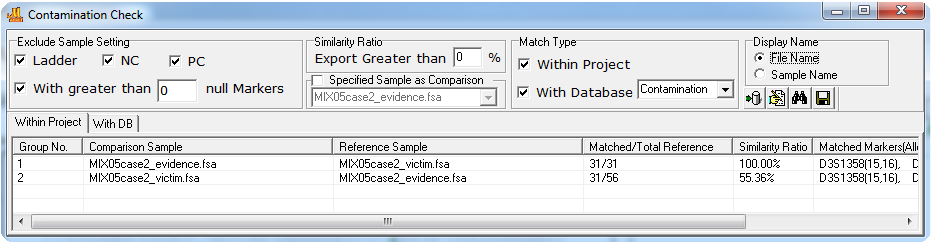
Figure 1: Profile-to-Profile check. In Group 1, 100% of the Reference sample alleles are included in the comparison sample. In Group 2, 55% of the alleles in the reference sample are included in the comparison sample. The last column of each row contains the shared genotype. The results can be saved as a .txt or .xlsx/.xls file from the disk icon.
Contamination Check Tool Results
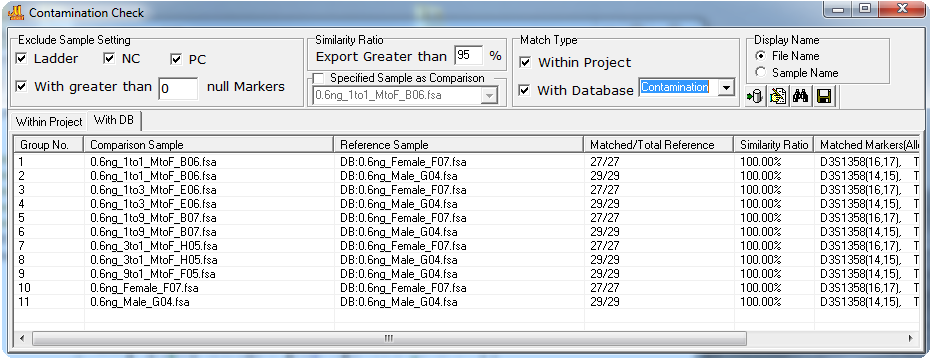
Figure 2: Profile-to-Staff or Elimination Database. The With DB tab displays the results comparing samples from the project with reference samples in the database. The single source file from the case and several other single source files were previously saved to the database to provide matches to the two samples of the project for this demonstration. The results indicate that 100% of the allele calls in MX05case2_victim sample are included in the comparison samples 1 and 2. Four profiles from the database (Reference Samples) have greater than 50% similarity to the Control Sample MX05case2_evidence.fsa of the current project. Note also that reference profiles retrieved from the database are designated with DB:filename in the Reference Sample column.
Calculation Details and Features of the Contamination Check Tool
The equation used to calculate the similarity ratio is:

The settings of the contamination tool may be customized per the lab SOP and include the following options:
- Include or exclude allelic ladders and controls
- Specify the number of Null Markers to allow before reporting a similarity ratio for a profile
- The minimum percent similarity
- Specify a file of interest (if this is left blank all samples in the project will be checked for contamination)
- Match type
- Within a project – (profile-to-profile)
- With DB (profiles-to-Database) - Select the database to screen (Contamination – staff member, Relationship testing or cell line databases)
- Display File name or Sample Name
- Icons to save genotypes to the contamination(staff) database, edit the database, refresh table after changing any parameters and save report tables
Application Notes:
Webinars:













