LINKED MIXTURE ANALYSIS FUNCTION & SEARCHABLE DATABASE
The Mixture Analysis Application is directly linked to the main analysis screen - avoiding the error-prone, human transfer of data from one software package to another. Repetitive calculations are automated, according to user-set analysis parameters.
See figures 1 – 4 (below) for an overview of the benefits of GeneMarkerHID mixture analysis tools
Click here if interested in Probablilistic Mixture Analysis with MaSTR™ Software
Short Tandem Repeat (STR) genotyping is instrumental in data analysis for a wide range of human identification applications. Forensic samples from mass disasters and crime scene investigations are mixtures rather than single source, which require additional analysis after obtaining DNA profiles (per SWGDAM Guidelines). GeneMarker®HID software performs consistent and accurate genotype determination, is compatible with commercial forensic chemistries and major CE genetic-analyzers (including ABI®PRISM, Applied Biosystems SeqStudio™, Promega Spectrum Compact CE System, or a rapid system such as RapidHIT™ and RapidHIT™ID). The embedded mixture analysis application distinguishes between mixtures of 2-contributors and more complex mixtures, automates repetitive calculations (Mixture-Ratios, Likelihood-ratios LRs, Probability-of-Inclusion, Probability-of-Exclusion, Random-Man-Not-Excluded), ranks the most likely genotype combinations (with or without single source reference samples) deducing contributor genotypes, and searches a database for a matching profile or potential relatives. The mixture analysis application uses allele frequency tables from USA populations, or easily imports other population allele frequencies to calculate LRs. This module was developed using recommendations of the DNA Commission of the International Society of Forensic Genetics (Gill et al., 2006) and methods of Clayton et al., 1998 and Gill et al., 1999.
Features of the Mixture Analysis Application include:
- Easy customization of analysis settings to meet the laboratory SOP
- Groups files by number of contributors
- Identifies contributor single source files
- Displays all possible allele combinations for contributors, color-coding those that meet all mixture analysis parameters
- Determines major and minor contributors
- Searches database for exact match or potential near relatives
Performs Repetitive Calculations:
- Major Mx ratio, Residual, Heterozygous Imbalance for each combination of alleles in two contributor mixtures
- Likelihood Ratios at each marker and combined LR in two contributor mixtures
- Probability of Inclusion and Probability of Exclusion for any mixture sample (two contributors or three or more)
Potential Mixture Samples are Identified Automatically in Main Analysis Screen
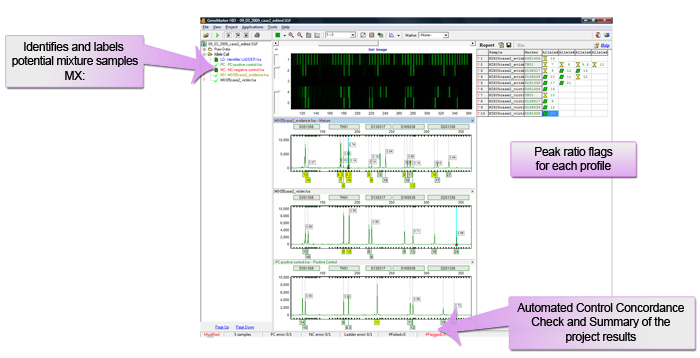
Figure 1: Main analysis screen showing flags for potential mixture samples.
Identify Potential Suspect without Suspect Reference Profile
Often, a suspect profile is not available. The mixture application deconvolutes the mixture. The report below displays the potential major contributor, which can be used to search laboratory, LDIS, SDIS or CODIS databases. All results are presented in report tables that may be saved or copy/pasted into existing documents.
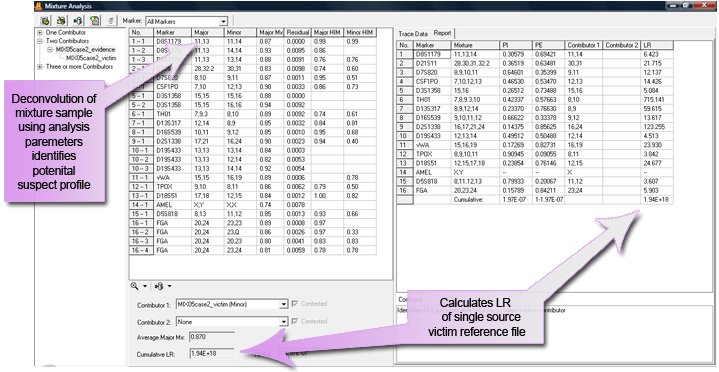
Figure 2: The Mixture Analysis application screen shows that mixtures can be deconvoluted and the LR calculated, even without a reference profile from a suspect.
Search database using major contributor potential suspect from deconvolution of mixture sample
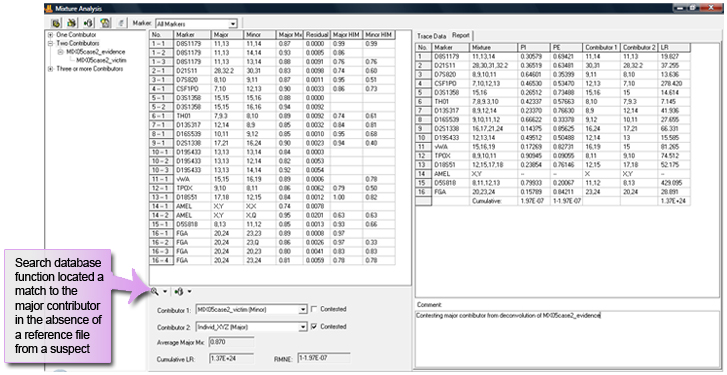
Figure 3: Click on the Search Database icon to locate matches to potential suspect profiles and calculate the mixture statistics.
Mixture Analysis with Two Single Source Reference Files

Figure 4: The Report table has an option to display the trace files of the mixture and all single source profiles that are contained with the mixture allele calls.
Webinars:
Reference Material:













