Annotation Tracks
The NextGENe Track Manager tool allows for import of variant annotation from external databases that can be automatically queried for use in both the mutation report (figure 1) and the variant comparison report. Tracks such as dbSNP, ClinVar, gnomAD, dbNSFP, and NHLBI GO Exome Sequencing Project (ESP) can be added to your projects, as well as custom databases. Once a track is imported for a given genome build, it can be automatically queried for every new project run against that genome or it can be queried manually in the NextGENe Viewer.
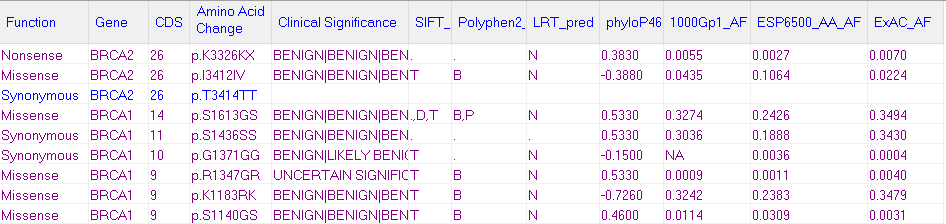
Figure 1: The NextGENe Mutation Report with annotation from multiple tracks.
There are four databases with specialized support. More information on some of these can be found in the ‘References' dialog of the NextGENe ‘Help' menu.
• dbNSFP (Database of Non-Synonymous Functional Prediction) - Functional Prediction, Conservation, and (when available) population frequency information is available for all non-synonymous SNPs

Table 1: List of sources in dbNSFP database
-
dbscSNV – in attached database with dbNSFP – functional annotations and ensemble
predictions for splice site substitutions
-
dbSNP/ClinVar
There is also a Custom Variant Track import option that can be used to import other databases not specifically listed above.
Querying Tracks in the Viewer
The "Query Reference Tracks" dialog can be used to import or update track information in an alignment project.
The available tracks for the applicable genome build for the project are listed (figure 2). Each track can be either selected so that it can be used in the reports for the project or can be deselected.
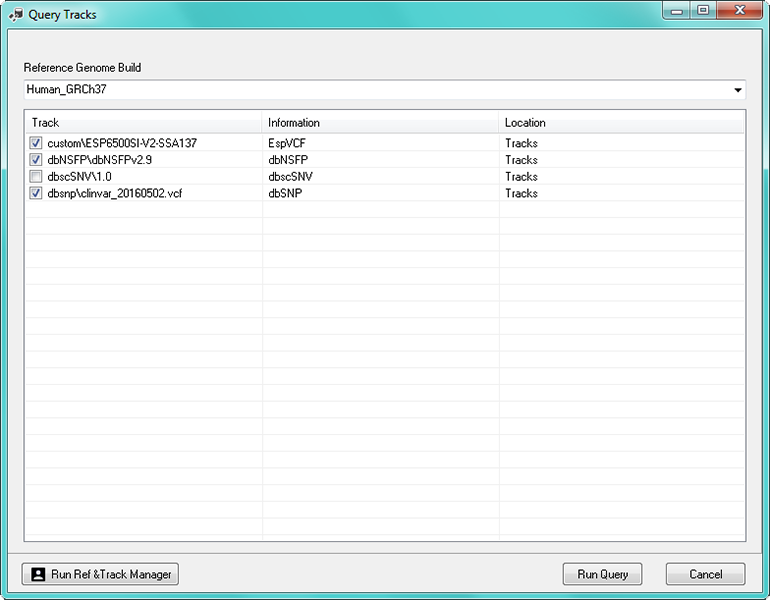
Figure 2: The "Query Reference Tracks" dialog.
Filtering using Tracks
For any track that is selected for a project, there are both display and filtering options available in the Variation Tracks Settings dialog (figure 3). You can select any track on the left side to define any filtering settings for track, and/or to click "Report Display" to choose which columns for the track to include in the report.

Figure 3: The Variation Tracks Settings dialog, dbNSFP track filter options shown.
Application Notes:
- Add Annotation Tracks from External Databases to NextGENe
- Finding Causative Mutation Candidates from NGS Sequencing
- Reducing error of NGS Sequencing data with NextGENe Condensation™ Tool
- Alignment of PGM PE reads with NextGENe software
- Processing Paired End Sequencing Data from the Ion PGM™ Sequencer with NextGENe® Software
- Processing Ion PGM™ Sequencing Data with NextGENe Software
- Processing CFTR Amplicon Data from the Ion PGM™ using NextGENe® Software
- Processing Ion AmpliSeq™ Cancer Panel Data using NextGENe® Software
- Amplicon Sequence Analysis of Sanger Sequences and Ion PGM data
- NextGENe Illumina SNP_Indel_Detection
- Structural Variant Analysis
- NextGENe software Variant Comparison Tool
- Mutation Confidence Scoring with NextGENe software
- NextGENe SNP & Indel Viewer
- Deep Sequence Analysis with NextGENe software
- Whole Genome Alignment with NextGENe software
- Sequence Capture Analysis with NextGENe software
- Paired end read merging with NextGENe software
- Sequence Analysis Using Barcode/Index Tags of Pooled Samples with NextGENe Software
- Using NextGENe software's Pipeline Automation Tool
- Demultiplexing Illumina® MiSeq™ Data
Webinars:
Pricing & Trial Version:
Reference Material:
Trademarks property of their respective owner













