Specialized RNA-Seq Analysis Tools
Transcriptome/RNA-seq, Expression and Alternative Splicing Analysis
Next Generation RNA sequencing is a powerful tool for examining the entire transcriptome, but alternative splicing analysis presents challenges when aligning RNA-seq data to a reference that are not encountered with alignment of DNA sequences. Short reads within exons are usually able to align normally, but challenges arise with reads that span exon-exon junction’s novel splicing events. NextGENe software provides solutions to these issues with analysis tools designed for RNA-Seq analysis of sequencing data from Illumina® iSeq, Miniseq, MiSeq, NextSeq, HiSeq, and NovaSeq systems, Ion Torrent Ion GeneStudio S5, PGM, and Proton systems as well as other platforms. Utilizing a novel algorithm not found in programs such as Q-PALMA, Supersplat, and TopHat, NextGENe software aligns reads belonging to annotated and novel transcripts while providing the added benefit of a highly graphical interface that does not require use of scripting or the command line. Variant detection, expression level analysis, and isoform identification are all performed in one analysis, with the NextGENe Viewer providing a high level of visualization for review and editing.
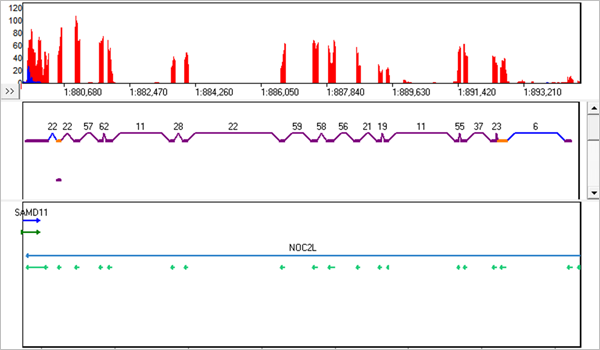
Figure 1: The new RNA-Seq Transcript View. The purple links and exons are known, the blue links are novel, and the orange exons are alternative splicing.
NextGENe software's approach to RNA-seq analysis takes advantage of a four-step proprietary algorithm to ensure that reads spanning exon junctions are aligned. The approach uses previously-known isoform splice sites but still allows for detection of novel transcripts. Reads are aligned to a pre-indexed reference using the NextGENe Whole Genome Alignment method. Transcripts are predicted based on the alignments. These predictions are compared to known transcripts and used to generate a transcriptome reference specific to the sample. Finally, the original reads are aligned to the discovered transcripts to ensure best alignment, recalling discovered transcripts and performing mutation detection of SNPs and short indels.
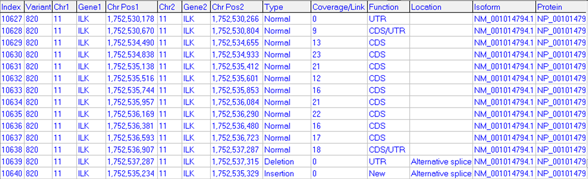
Figure 2: Transcript report
The transcript report provides information about detected exons such as location, coverage/link number (reads spanning an exon junction), type of variant (insertion, deletion, new/novel, or normal exon), function, location type (alternative splice site, exon skipping, etc.), and isoform/protein information. All reports are highly customizable with many different filter and display options, and expression levels are reported in the NextGENe Expression Report.
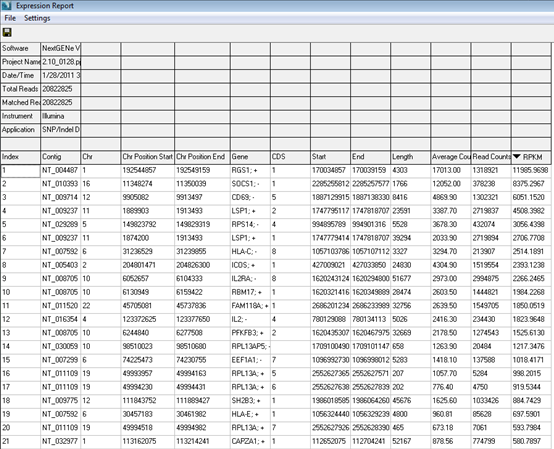
Figure 3: Expression report sorted by RPKM.
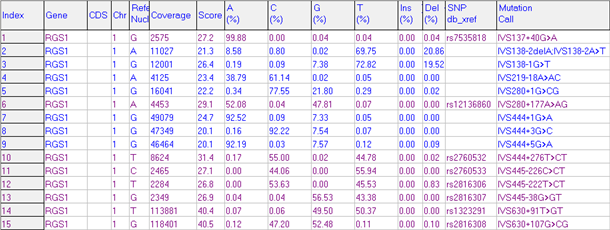
Figure 4: Mutation Report
Application Notes:
- Ion PGM RNA-Seq Analysis Application Note
- RNA-Seq Analysis with NextGENe Software
- Transcriptome Analysis Application Note
- Using NextGENe software's Pipeline Automation
Webinars:
Pricing & Trial Version:
Reference Material:
Trademarks property of their respective owner













