Family/Trio and Multi-Sample Variant Comparison
The NextGENe Variant Comparison Tool can be used to compare variants detected across samples, including trio analysis, tumor-normal pairs and other comparisons. This is a valuable tool to quickly sort through thousands of mutation calls in order to find a few dozen (or even fewer) candidate mutations that can be confirmed with Sanger sequencing and assessed for their phenotypic impact. Information from imported tracks, such as dbNSFP, dbSNP, gnomAD and any custom tracks, can be displayed in the comparison tool and can also be used for filtering the variants included in the report.
There are multiple options available for creating the comparison:
• Based on sample relationships, phenotypes and the applicable inheritance pattern
• Based on specified mutation types, or similarity or differences between the samples
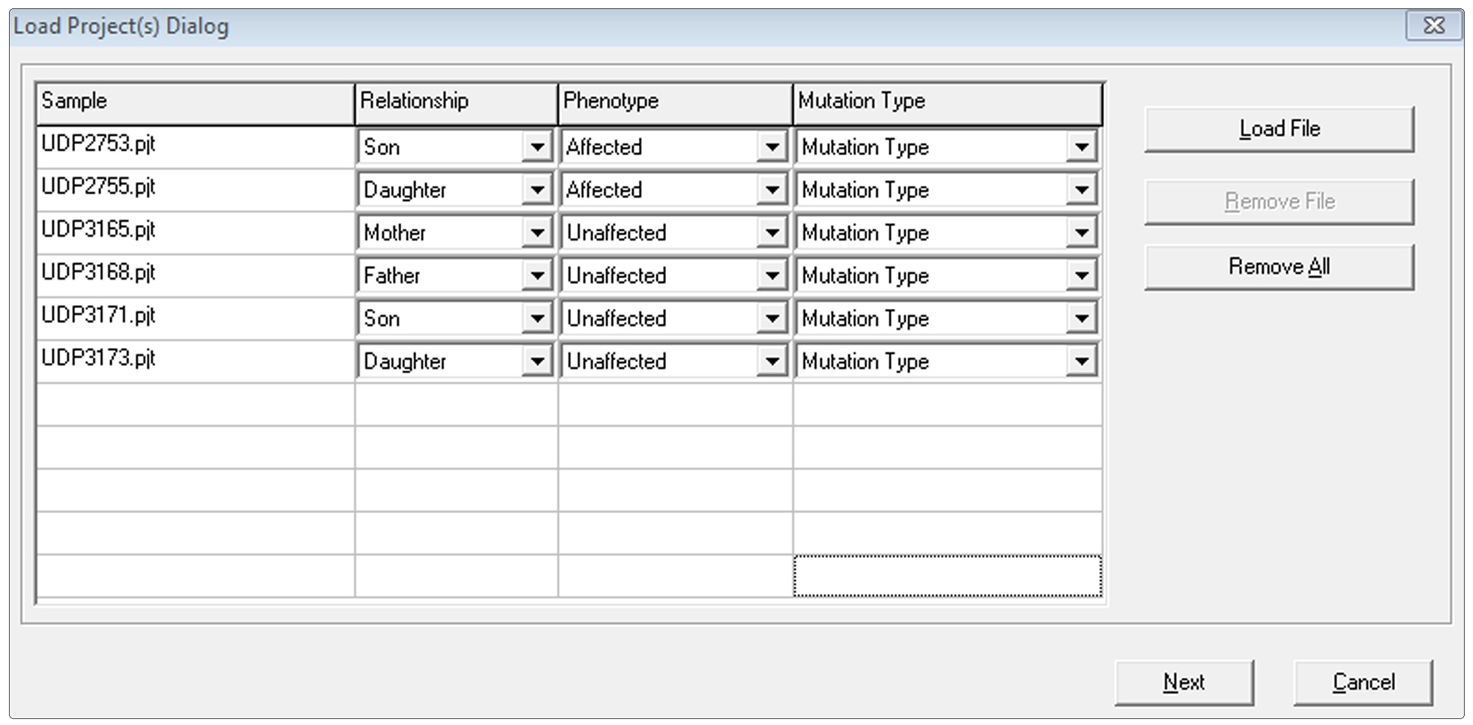
Figure 1: Specify relationships and phenotypes for each sample to compare variants based on an inheritance pattern. The applicable inheritance pattern can be selected in the next dialog.
The resulting comparison view includes a report table as well as visualization for each sample side by side. The table and visualization are linked - double-clicking on any variant in the report directly loads the visualization for that position in all samples.
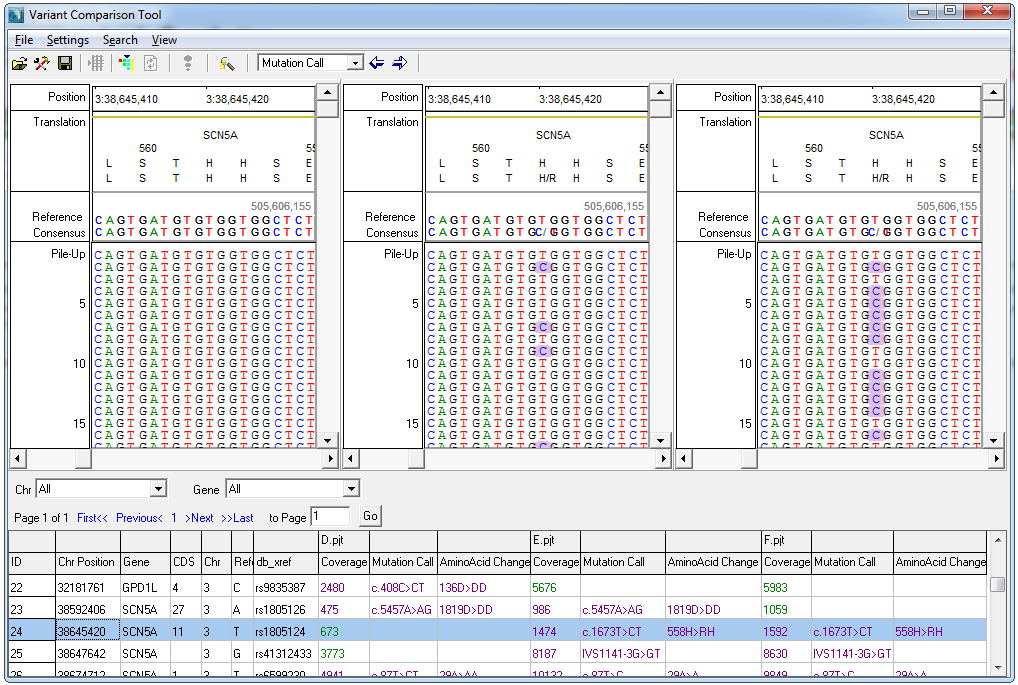
Figure 2: The Variant Comparison tool comparing 3 samples. A report table and alignment view for each sample is displayed.
Application Notes:
- Finding Causative Mutation Candidates from NGS Sequencing
- Reducing error of NGS Sequencing data with NextGENe Condensation™ Tool
- Alignment of PGM PE reads with NextGENe software
- Processing Paired End Sequencing Data from the Ion PGM™ Sequencer with NextGENe® Software
- Processing Ion PGM™ Sequencing Data with NextGENe Software
- Processing CFTR Amplicon Data from the Ion PGM™ using NextGENe® Software
- Processing Ion AmpliSeq™ Cancer Panel Data using NextGENe® Software
- Amplicon Sequence Analysis of Sanger Sequences and Ion PGM data
- NextGENe Illumina SNP_Indel_Detection
- Structural Variant Analysis
- NextGENe software Variant Comparison Tool
- Mutation Confidence Scoring with NextGENe software
- NextGENe SNP & Indel Viewer
- Deep Sequence Analysis with NextGENe software
- Whole Genome Alignment with NextGENe software
- Sequence Capture Analysis with NextGENe software
- Paired end read merging with NextGENe software
- Sequence Analysis Using Barcode/Index Tags of Pooled Samples with NextGENe Software
- Using NextGENe software's Pipeline Automation Tool
- Demultiplexing Illumina® MiSeq™ Data
Webinars:
Pricing & Trial Version:
Reference Material:
Trademarks property of their respective owner













