Using NIST Genome in a Bottle Reference Samples to Validate an NGS Analysis Pipeline with NextGENe® and Geneticist Assistant® Software
The Genome in a Bottle (GIAB) Consortium hosted by NIST provides reference materials to facilitate the adaptation of next generation sequencing tests into clinical practice. The reference materials, based on NA12878 DNA from Coriell, are a valuable tool for validating an NGS pipeline as the pipeline results can be compared with the highly confident small variant (SNP and indel) calls from GIAB.
NextGENe and Geneticist Assistant software provide a complete user-friendly analysis pipeline for NGS data from the raw data off of the sequencer to curated variant calls and variant interpretation. GIAB samples can be used to validate the optimal analysis and reporting settings by specifically evaluating high confidence variant calls. NextGENe software offers flexible variant calling and reporting settings for filtering variant calls and Geneticist Assistant NGS Workbench can be used to compare NextGENe calls with a VCF of the known calls for the sample from GIAB. In this way settings can be adjusted to eliminate potential false negatives while minimizing false positives.
NextGENe Mutation Report Settings can be configured to report every position within the GIAB VCF in order to easily identify false negatives.
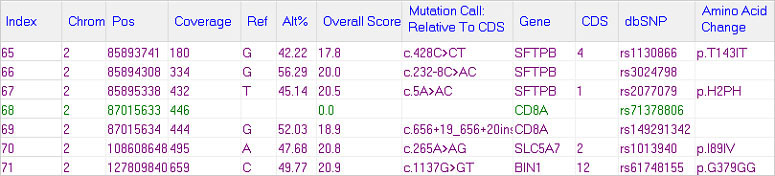
Figure 1: In the NextGENe Mutation Report variants that are included in the GIAB high confidence variant VCF but not called in the sample (potential false negatives) are shown with green text and the Mutation Call column is blank.
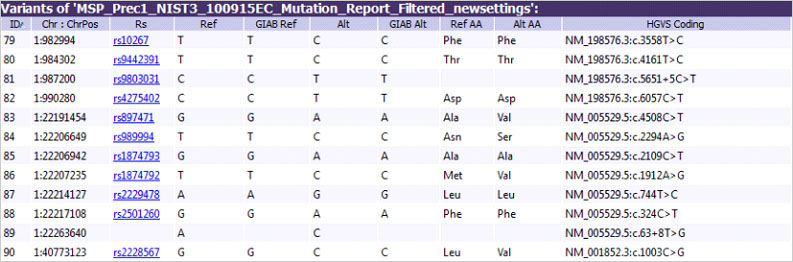
Figure 2: In Geneticist Assistant NGS Workbench, annotation from the GIAB VCF such as the Ref and Alt Alleles can be added to the variant table allowing viewing of information for all variants in the sample in a single view.
Application Notes:
Webinars:
Trademarks property of their respective owner













