Prediction and Rare Disease Discovery
In collaboration with researchers at the NIH, SoftGenetics has developed a biologist and geneticist-friendly tool designed to speed up and simplify the discovery of causative mutations. Sequencing the exomes of a small family or several individuals with similar phenotypes can result in a list of hundreds of thousands of possible mutations.
The NextGENe Variant Comparison and prediction database tool makes it easy to narrow down the found variation list based on several levels of criteria:
- Expected genotype - including control samples and inheritance patterns
- Annotated gene information - exons, coding sequences, splice sites, etc
- Database information -dbSNP, NHLBI GO Exome Sequencing Project (ESP), custom databases
- Functional Prediction - dbNSFP (including 1000 genomes frequencies, PolyPhen2, SIFT, PhyloP and more)
- Type of mutation - Indels or Substitutions (Silent, Missense, or Nonsense)
- Inclusive and Exclusive region of interest (ROI) filtering
- Mutation confidence score filtering
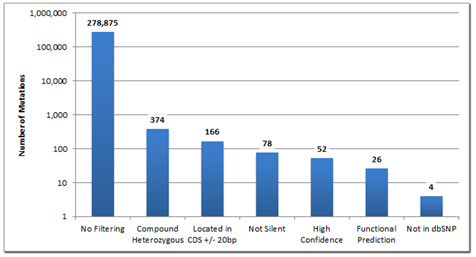
Figure 1: In this family of 6, with two affected children, the exomes of the entire family- mother, father, two unaffected and the two affected children were sequenced. The nearly 280,000 shared variants were quickly reduced by the NextGENe variant comparison tool allowing geneticists to quickly ascertain potential causative mutations.
Application Notes:
- Finding Causative Mutation Candidates from NGS Sequencing
- Processing Ion PGM™ Sequencing Data with NextGENe Software
- Processing Ion AmpliSeq™ Cancer Panel Data using NextGENe® Software
- NextGENe Illumina SNP_Indel_Detection
- Structural Variant Analysis
- NextGENe software Variant Comparison Tool
- Mutation Confidence Scoring with NextGENe software
- NextGENe SNP & Indel Viewer
- Deep Sequence Analysis with NextGENe software
- Whole Genome Alignment with NextGENe software
- Sequence Capture Analysis with NextGENe software
- Using NextGENe software's Pipeline Automation Tool
Webinars:
Pricing & Trial Version:
Reference Material:
Trademarks property of their respective owner













