SNP/Indel Detection of Next Generation Sequence (NGS) Reads
Next Generation Sequencing has made it possible to rapidly examine individual genomes and compare genetic sequences across multiple genomes to detect variants. NextGENe® software has been developed specifically for use by biologists performing whole genome sequencing (WGS), whole exome sequencing (WES), or targeted resequencing projects containing single nucleotide polymorphisms (SNPs), Insertions/deletions (indels), and large structural DNA rearrangements from Next Generation Sequencing data produced by Illumina® iSeq, Miniseq, MiSeq, NextSeq, HiSeq, and NovaSeq systems, Ion Torrent Ion GeneStudio S5, PGM, and Proton systems as well as other platforms. NextGENe software combines patented algorithms and analysis tools designed to run on a Windows® operating system, eliminating the need for scripting and other bioinformatics support needed in programs such as CLC Genomics Workbench, Lasergene's SeqMan Pro, as well as academic software such as MAQ & SOAP, Top Hat, BWA & Bowtie. In addition to powerful analysis tools, NextGENe software offers automated and advanced creation of custom reports and quality metrics as well as a biologist friendly annotated whole genome browser/viewer for review, filtering, editing, and commenting.
Instrument specific SNP/INDEL analysis:
Illumina System Analysis
Ion Torrent Sequencing Platforms
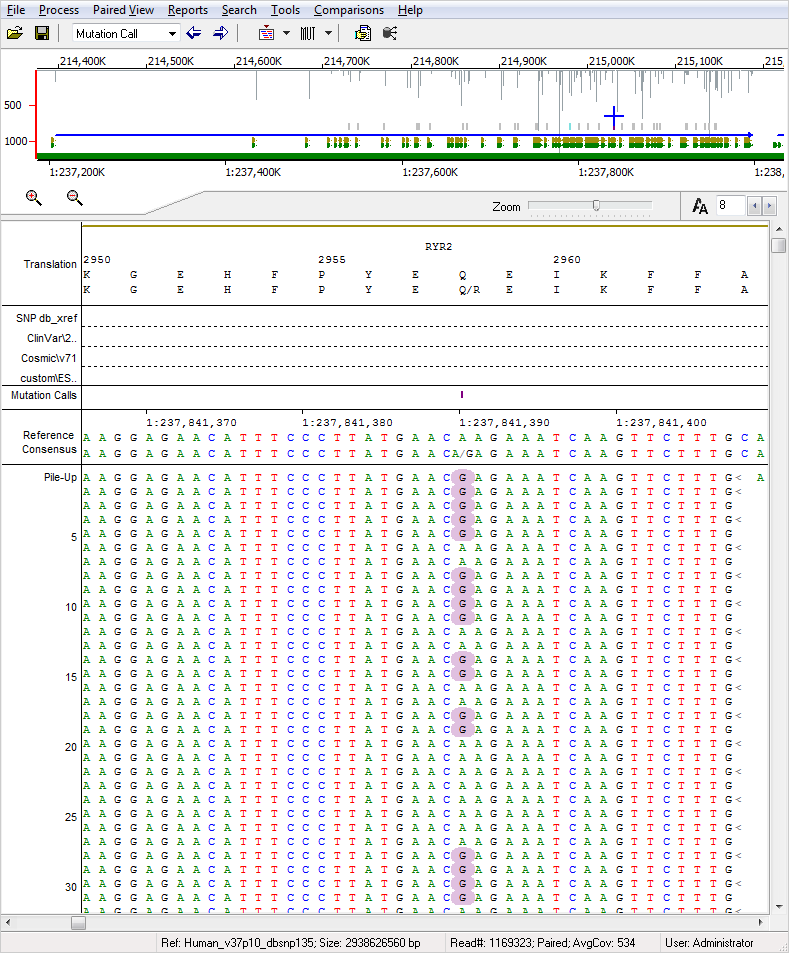
Figure 1: NextGENe Viewer software displaying a SNP with purple background. NextGENe software features up to 99% accuracy, with the ability to detect indels up to the read length, and rapid review of DNA variants, nucleotides, and amino acids. Alignment and variant detection can be completed using a whole genome reference or targeted regions using GenBank or fasta files. Upon completion of a sequence alignment project, interactive mutation reports are automatically generated providing detailed variant and annotation information. Reports also include a host of filtering options, variant confidence scoring, multiple analysis comparison capabilities, hyperlinks to dbSNP, and additional tracks with causative prediction through dbNSFP, as well as custom database import including ExAC, gnomAD, and NHLBI GO Exome Sequencing Project (ESP).
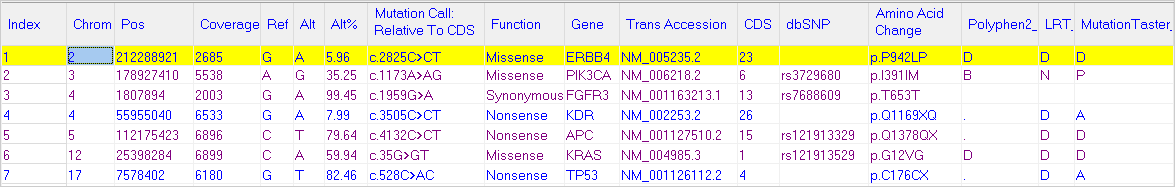
Figure 2: Mutation report with custom track information, including 1000 Genomes and dbNSFP.
Database information can easily be imported in NextGENe software. Updated dbSNP information is included in the human genome reference provided by SoftGenetics as well as select GenBank files downloaded from NCBI, providing hyperlinked access from the mutation report. Additionally, the NextGENe Track Manager Tool enables users to quickly import the most recent data from dbNSFP or custom databases. Information from these database can be used for display and/or filtering the Mutation Report.
NextGENe software also has an enhanced processing optimization providing 60% faster processing time. NextGENe software has the capability of creating an automatic link between NextGENe and Geneticist Assistant Workbench software, creating a seamless Analysis/QC/Variant Interpretation/Reporting pipeline.
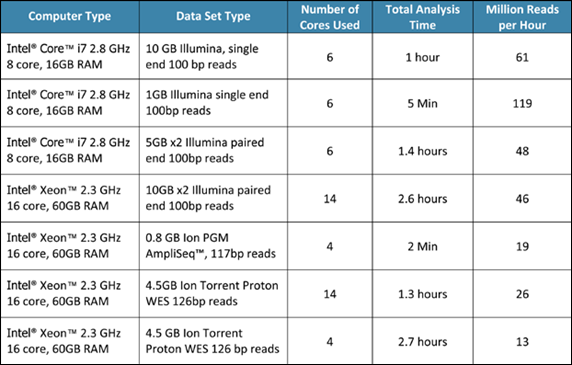
Performance Examples on various datasets using NextGENe Software:
Application Notes:
- NextGENe® Software Analysis Using the NEBNext Direct® Cancer Hotspot Panel
- Highly Sensitive Somatic Mutation Detection from Biopsy Samples with Deep Sequencing using NextGENe® Software
Webinars:
Pricing & Trial Version:
Reference Material:
Trademarks property of their respective owner













