Structural Variation Detection using NextGENe software
Over 68,000 structural variants (SVs) have been reported in the human genome, including deletions, duplications/insertions, and translocations such as gene fusions (1). Alone, there are more than 2,200 catalogued gene fusions (2) making genomic disorders an important area of research (3).
There are several techniques available for detecting SVs. Microarrays, for one, can be useful for detecting copy number variation but are limited to detecting events relative to specific design probes (4). Next Generation Sequencing can produce higher resolution results and enable the identification of novel events as well as the simultaneous detection of SNPs and small indels.
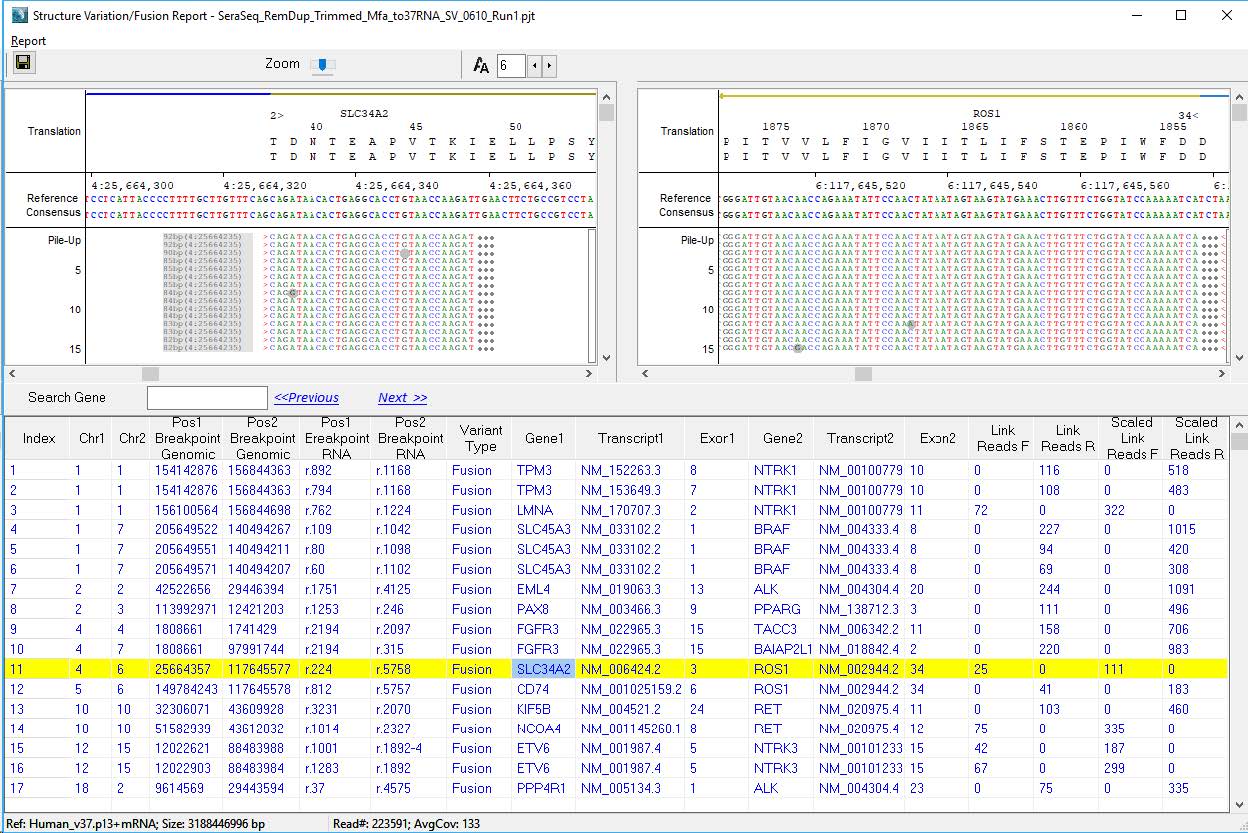
Figure 1: NextGENe Viewer’s Structural Variation Report is an interactive tool for displaying both breakpoints of each detected SVs.
NextGENe software can align reads simultaneously to genomic and transcriptomic references. Reads from RNA samples can align across exon junctions more easily, enabling higher sensitivity for the detection of breakpoints. Next generation sequencing reads from instrument manufactures such as Illumina® can be analyzed by NextGENe software for the detection of SVs. Reads with a high level of mismatch at an end are split into two reads and remapped to the DNA/RNA reference. These Link Reads identify the breakpoints of structural variations including gene fusions.
NextGENe software includes templates to easily analyze gene fusion samples, streamlining the analysis of many SVs.
The Seraseq Fusion RNA Mix v2 from SeraCare Life Sciences, Inc. includes many gene fusions, like TPM3-NTRK1, LMNA-NTRK1, SLC45A3-BRAF, EML4-ALK, PAX8-PPARG, FGFR3-TACC3, FGFR3-BAIAP2L1, SLC34A2-ROS1, CD74-ROS1, KIF5B-RET, NCOA4-RET, and ETV6-NTRK3.
A sample was run on an Illumina MiSeq instrument and produced about 255K 2x150bps reads. Trimming of adapters yielded reads averaging 112bp. After utilizing the Overlap Merger tool, 90.5% of the pairs were merged into single reads with an average length of 140bp, extending the average length by about 30bp, (see Figure 4).
NextGENe software was run on a standard Windows Desktop computer. 223K (96.4%) of these reads are aligned to the DNA/RNA reference genome in 3 minutes and Structural Variation detection is completed in 8 additional minutes by NextGENe software. Reports in TXT, VCF and PDF formats are automatically generated.
The final project can be opened in the NextGENe Viewer software, which offers an interactive tool for viewing the SVs and producing additional reports (see Figure 1). Reads that are split and mapped across breakpoints are called Link Reads, and about 2K of these reads are mapping to all 12 expected fusion events in the SeraCare fusion controls.
Application Notes:
- Structural Variant Detection Application Note
- Analysis of Advanta RNA Fusions Assay Samples Application Note
Webinars:
Pricing & Trial Version:
Reference Material:
Trademarks property of their respective owner













