

Hypervariable regions of the genome contain extreme levels of polymorphism and genetic variation. Hypervariable regions include the D-loop of mitochondrial DNA, the human leukocyte antigen (HLA) gene family of the major histocompatibility complex, and the RNA of retroviruses such as the human immunodeficiency virus (HIV) and the hepatitis C virus (HCV). Sanger sequencing projects involving a high level of variability demand fully automated calls of single nucleotide polymorphisms (SNPs), low-frequency variants (somatic, mosaic, heteroplasmic) and homozygous and heterozygous insertions, deletions, and duplications (indels) to identify variations between strains. Mutation Surveyor software's patented “anti-correlation” technology performs a physical trace-to-trace comparison of hypervariable sequences to a reference trace in order to detect sequence variants. The software is perfect for the analysis of hypervariable regions, offering accuracy greater than 99% in the bi-directional analysis mode and sensitivity down to 5% of the primary peak using Phred 20 bi-directional sequence data.
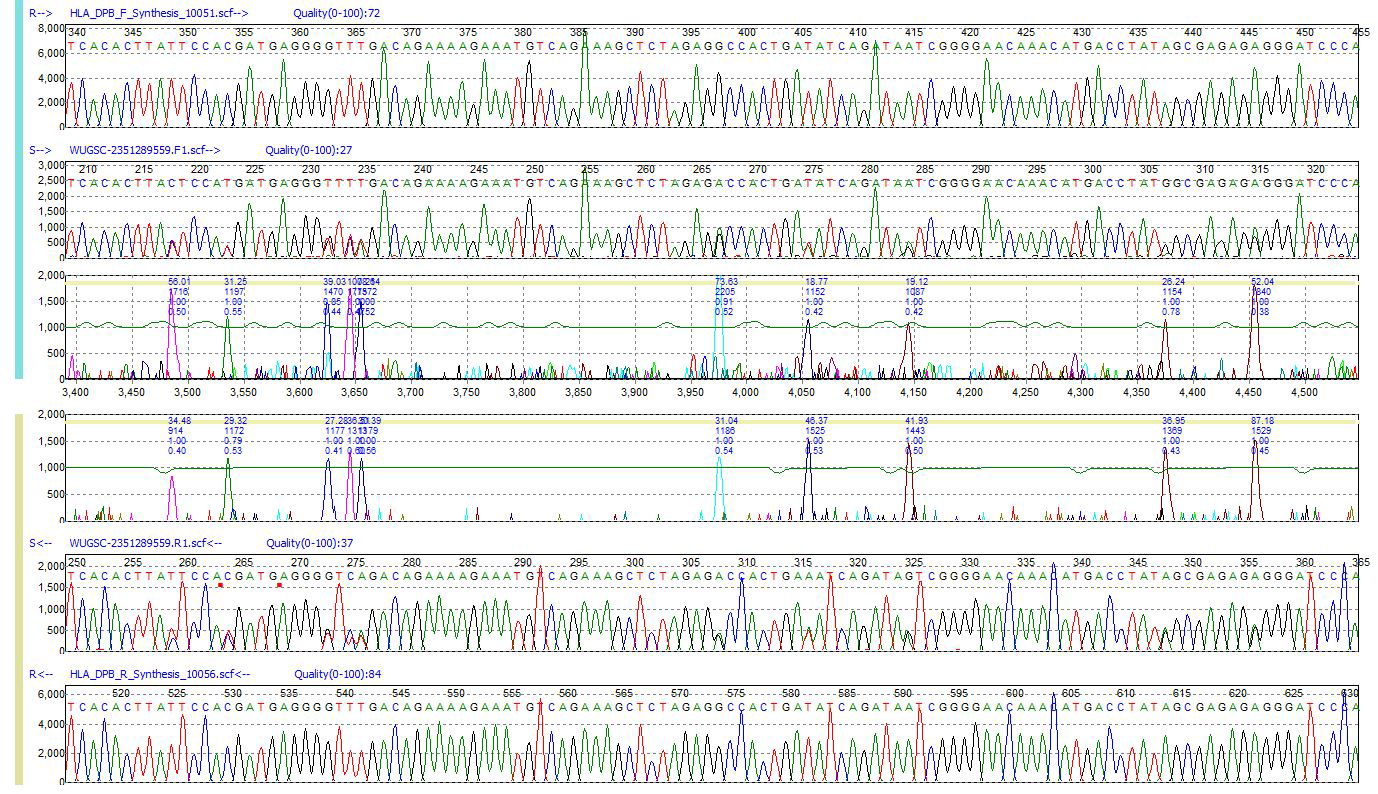
Figure 1: HLA-DPB1 sample showing a high frequency of mutations when compared to the reference trace. The mutation electropherograms display mutations based on the level of correlation between sample and reference trace.
Application Notes:
Webinar
Trademarks property of their respective owner