Variant Knowledge Base with External Annotation
Mutation Surveyor software includes a variant knowledge database, which allows users to indicate false-positives and other artifacts, as well as positions of significance by chromosomal position. Users can easily flag common artifacts and variants with the click of a mouse, adding the entries to the custom database to be remembered and queried for future projects. Also included in the knowledge base is a Reference & Track Manager tool that provides annotated results in the Graphic Analysis Display and some reporting options. Annotation from external databases such as dbSNP, ClinVar, gnomAD, and dbNSFP – including causative prediction – as well as custom database information can be added to mutation projects to highlight positions of significance.
Variant Positions from Imported External Variant Databases are displayed as Green Tick Marks in the Variation Tracks Pane of the Graphical Analysis Display

Figure 1: Track annotation pane featuring imported database information and chromosome location. Each green tick mark represents database annotation at that position.
The Reference & Track Manager tool in Mutation Surveyor software allows users to import variant annotation from external databases that can be automatically queried for use in the Graphical Analysis Display and some variant reporting options. Annotation databases such as dbSNP, 1000 Genomes, and dbNSFP can be added to projects. Once a database is imported for a given preloaded reference, it can be automatically queried for new projects.
Annotations from Imported External Databases are Displayed in the Annotation Column of the Mutation Table in the Graphical Analysis Display
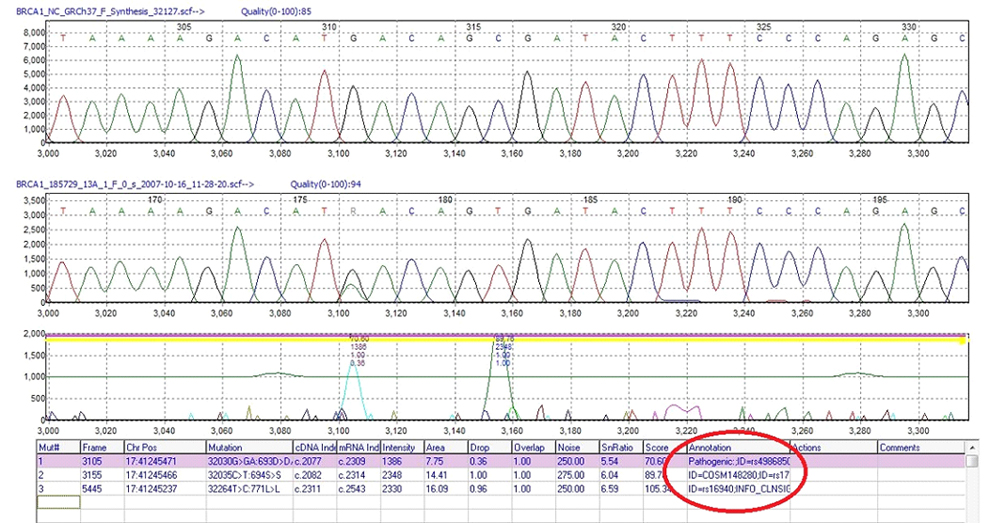
Figure 2: Mutation table in Graphical Analysis Display showing chromosome position and annotation column with database information such as dbSNP IDs.
The Variant Knowledge Base of Mutation Surveyor software allows users to track common edits made during variant analysis. Predictable errors that occur near the beginning and end of sequencing runs can easily be added to the database as sequencing artifacts for automatic removal from future mutation projects. Alternatively, common false positives due to sequencing chemistries may be labeled as such and added to the database for further review.
Deleted Variant Calls Known to be Artifacts and False Positives Can be Added to the User Knowledge Database for Automatic Flagging and Deletion in Future Projects
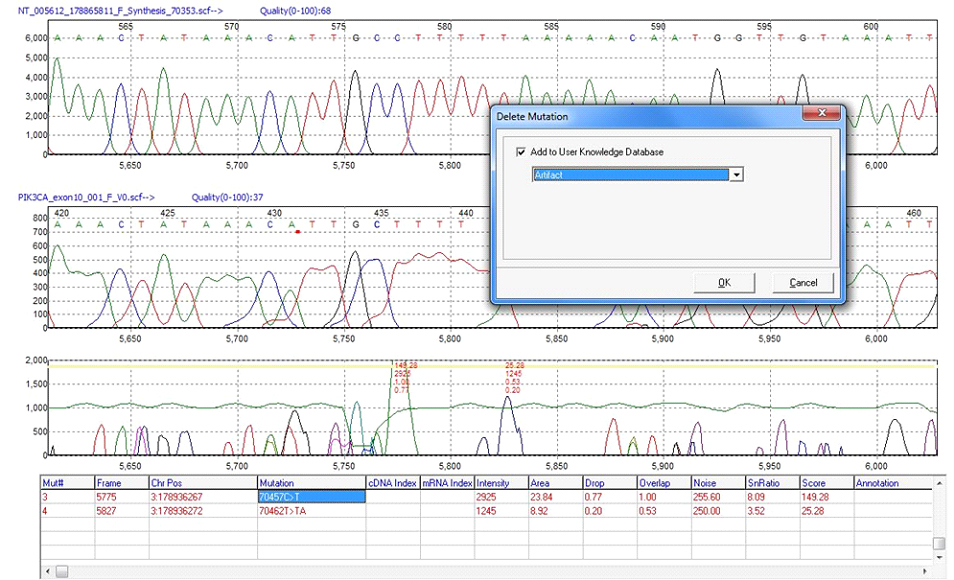
Figure 3: Deletions can be added to the User Knowledge Database and queried in future Mutation Surveyor projects.
Application Notes:
Track Manager Manual:
Use the Reference & Track Manager tool to import and manage preloaded references and data from any public or proprietary variant database.
Webinars:
- Application of Annotation from External Databases to Mutation Surveyor Projects
- Mutation Surveyor User Knowledge Database with External Annotation
- Creating a User Knowledge Database of Known Artifacts / False Positives
Trademarks property of their respective owner













