Heterozygous Insertion and Deletion (Indel) Detection and Deconvolution in Sanger Sequencing Traces
Mutation Surveyor software employs a multi-step alignment algorithm to detect homozygous and heterozygous deletions, insertions, and duplications (indels) from Sanger Sequencing traces. Included in the software is a unique and interactive Heterozygous Indel Detection tool not found in other sequencing software such as Thermo Fisher Scientific SeqScape®, Variant Reporter™, and Minor Variant Finder; DNAStar's SeqMan Pro™; Gene Code's Sequencher®; or JSI Medical Systems' SEQUENCE PILOT. This tool automatically detects and deconvolutes heterozygous indels into wild-type and novel alleles, eliminating the need for tedious and error-prone manual deconvolution. Mutation Surveyor software is able to detect heterozygous indels with sensitivity at 20% contribution from the minor allele by monitoring the migration time changes between the sample and reference trace. After alignment of the sample trace to the reference trace the novel/mutant sequence is deduced by subtracting the reference from the sample sequence. The mutant trace is automatically shifted to realign with the reference sequence, providing the location of the variant in an interactive display window.
Automatic Deconvolution of Heterozygous Indels in Heterozygous Indel Detection Tool
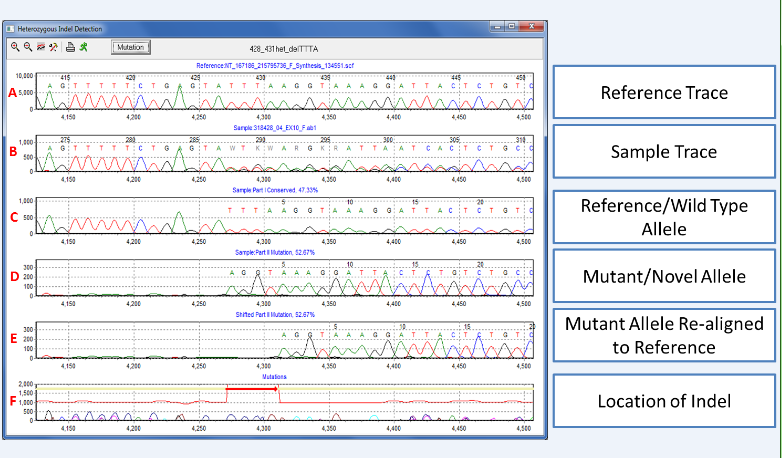
Figure 1: A 4bp heterozygous deletion of TTTA is detected in the Heterozygous Indel Detection tool. Trace A is the reference and trace B is the sample trace containing a mixture of alleles. The sample trace is automatically deconvoluted to display the normal/wild-type allele in trace C and the novel/mutant allele in trace D. The mutant allele is then shifted to realign to the reference in trace E and the location depicted in trace F. The mutant percentage, displayed above each allele, is calculated by taking the ratio of the mutant intensity over the mutant plus normal intensity (total).
When a heterozygous indel is detected during the alignment process, Mutation Surveyor software will automatically deconvolute the mixture to determine the location of the event. For further review, the Heterozygous Indel Detection tool can be opened to examine and edit the indel. Trace A is the reference trace and trace B is the sample trace containing the mixture of two alleles. The sample trace is automatically deconvoluted into wild-type allele in trace C and novel/mutant allele in trace D. Finally, the tool shifts the mutant trace to align to the reference in part E, providing a clear location of the event in trace F. The tool is highly interactive, allowing users to zoom in and out of the display window, and print images at high resolution.
Mutation Surveyor software’s alignment algorithm compares the mobility of the sample trace to the reference trace to detect insertion, duplication, and deletion events. When a heterozygous indel is detected, a horizontal, brown bar is placed at the location of the event in the Mutation Electropherogram. The event can then be viewed in the Heterozygous Indel Detection tool by double-clicking on the mutation call in the Mutation Table, or by opening the tool from the icon at the top of the screen. The software is able to regularly and accurately detect various sizes of indels, with recent studies showing accurate detection of 3 to 30 base pair indels.
Graphical Analysis Display Indicates Start Site of Heterozygous Indel Event with Brown Horizontal Bar in Mutation Electropherogram

Figure 2: A heterozygous indel displayed in the Mutation Surveyor Graphical Analysis Display (GAD). The software places a horizontal brown bar at the start site location of the event for easy identification.
Application Notes:
Webinars:
Trademarks property of their respective owner













