Contig Reference Assembly and Variant Review in Mutation Surveyor
Following Sanger sequencing trace alignment and automatic detection of variants, Mutation Surveyor software provides a unique contig reference assembly and variant viewer which would negate the need for additional software packages such as Thermo Fisher Scientific SeqScape®; Gene Code's Sequencher®; or JSI Medical Systems' SEQUENCE PILOT. The Project Reviewer Report of Mutation Surveyor provides a versatile way of grouping and viewing sample files that are aligned to the same reference, including an overlapping view of individual contigs, an overlapping view of all contigs mapping to the same GenBank file, or an overlapping view of sample files associated with a patient identifier. Viewing and editing variants is also simplified in the Project Reviewer Report, which color codes variants and provides multiple ways to view sequence traces. The viewer is highly interactive, offering linked navigation between four data windows, including links to the sample and mutation electropherogram windows, and mutation report for quick and easy confirmation of variants such as single nucleotide polymorphisms (SNPs), low-frequency variants (somatic, mosaic, heteroplasmic), and homozygous and heterozygous insertion and deletion (indel) events.
Interactive Viewer Features Four Linked Data Windows
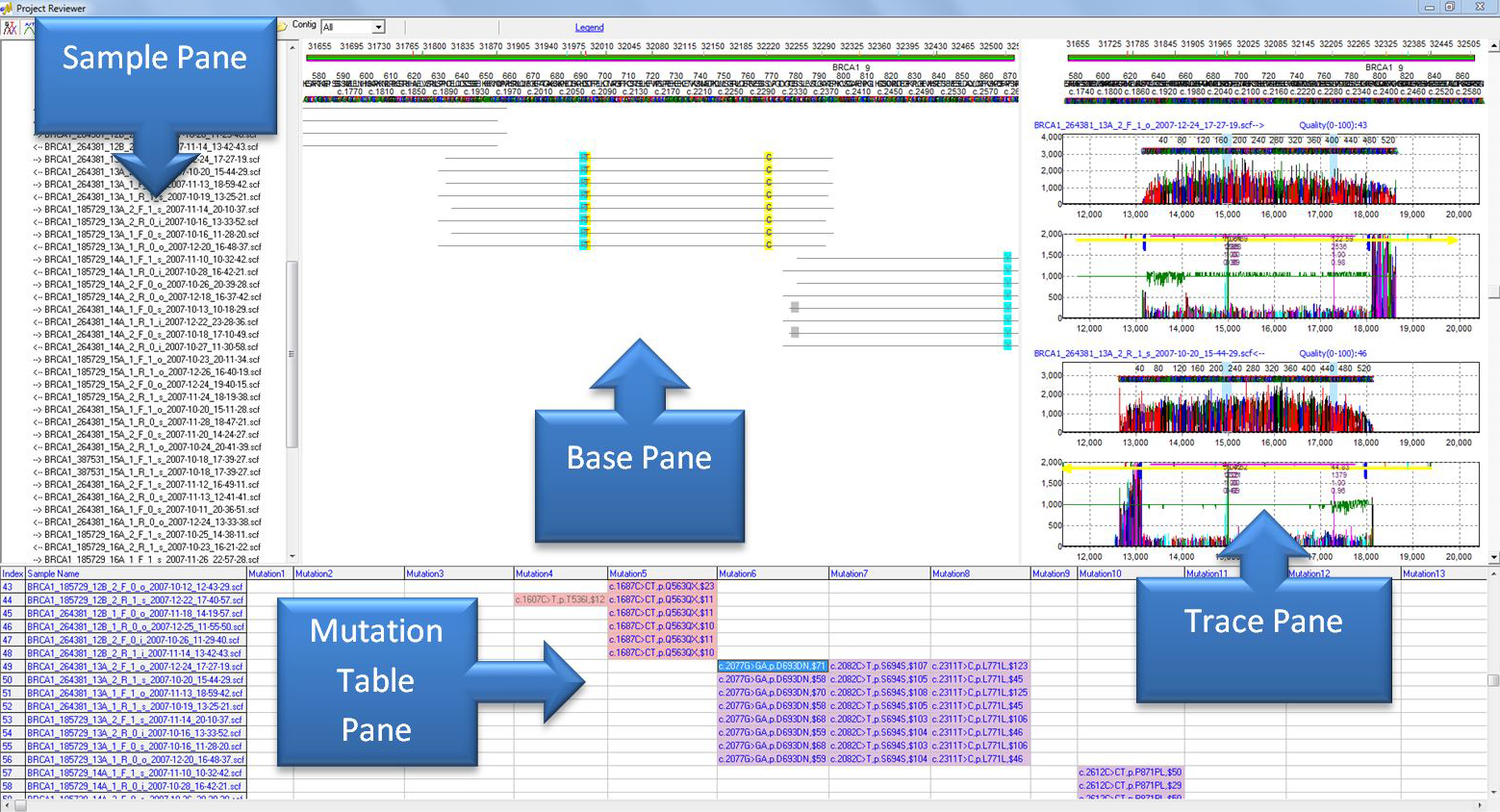
Figure 1: The Project Reviewer Report of Mutation Surveyor provides four interactive and hyperlinked views of Sanger sequencing traces in a mutation project. Double-clicking on a sequence trace in the sample pane will update the base and trace panes with the appropriate sequence. The base pane provides color-coded variants and the sample alignment to the reference as well as its overlap to other sequence traces. The trace pane provides the sample and mutation electropherograms for each sample trace. Variants are listed in the mutation pane at the bottom of the viewer, providing a convenient way to confirm or edit mutations with the click of a mouse. Each pane is collapsible, allowing the user to add or remove panes and readjust window sizes.
Review and Edit Mutation Projects
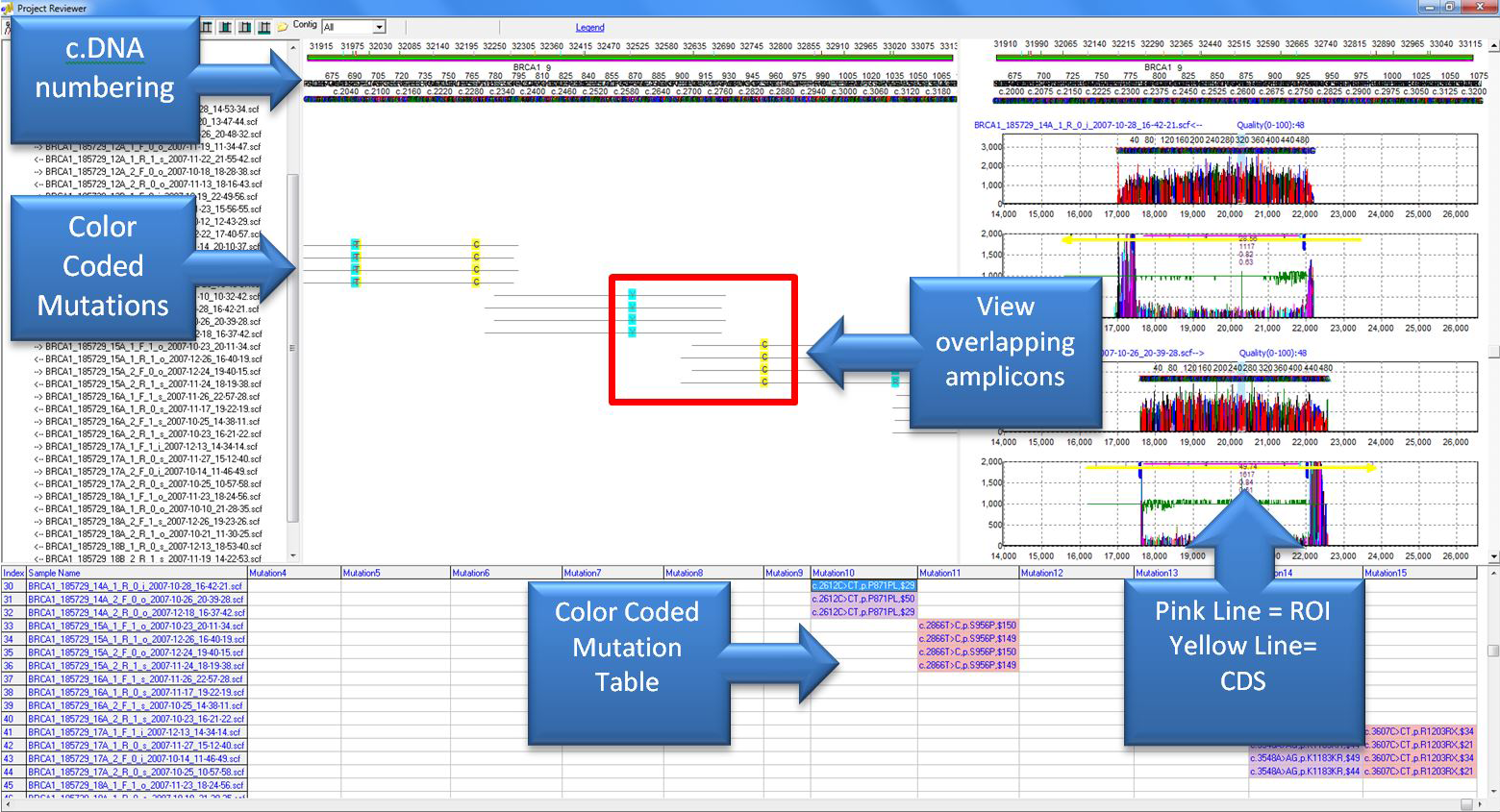
Figure 2: Overlapping amplicons are displayed in the base pane along with color-coded mutation calls. Annotation is provided by the reference GenBank or SEQ file loaded into the project. Annotation includes c.DNA numbering at the top of the report, regions of interest and CDS identifiers in the electropherograms, and mutation calls in the mutation table. The color-coded mutation table can display variants relative to genomic, CDS, mRNA, HGVS, or forensic (SWGDAM) annotation. Mutations can be edited or confirmed in the trace pane and mutation table pane.
Group Samples by Patient, Gene, or Contig

Figure 3: The Project Reviewer Report shows replicate samples for three different individuals aligning to the same reference sequence. The sample files were grouped by a patient identifier located in the sample file names. Both the mutation table pane and the base pane display variants detected in the same positions in all three patients. Multiple electropherograms can be seen simultaneously in the trace pane in order to quickly compare mutation calls between samples.
Webinars:
Trademarks property of their respective owner













