Haplotype Analysis and Automated Phase Assignment Pedigrees from Familial DNA STR Fragment Data
Familial DNA fragment data is used for haplotype analysis in areas such as genetic disorder research and preimplantation studies. Importing data from ABI®PRISM, Applied Biosystems® SeqStudio, or Promega Spectrum Compact CE systems genetic analyzers into GeneMarker software provides user-friendly, accurate genotyping with a linked Haplotype Analysis application, avoiding potential errors during data transfer from one software to another. GeneMarker software also combines allele call information from multiple PCR reactions for each individual (an alternative to Cyrillic©). Pedigrees are automatically drawn and allele calls of children and parents are used to assign a first-order approximation phase. Whenever the alleles are informative for phase assignment a pattern/color bar is assigned to: indicate the most probable phase, flag allele conflicts, detect potential uniparental disomy, and to allow analysts to review, edit for cross-over, as well as save and print pedigrees.
Features of GeneMarker® Software's Haplotype Analysis Application:
- Follows Bennett et al. nomenclature for pedigrees
- X-linked and autosomal pedigree formats based on parent/child(ren) allele calls
- Edit family and individual information
- Displays markers, allele calls, personal information in pedigree
- Control ordering of markers in pedigree by customizing panels
- Automatically makes first order phase assignment
- Edit capability for reassigning phase or edit crossover
- Save pedigree and re-open for adding data and editing
- Allele conflicts flagged; detects potential uniparental disom
Results X-Linked (left) and Autosomal Linkage (right) Examples
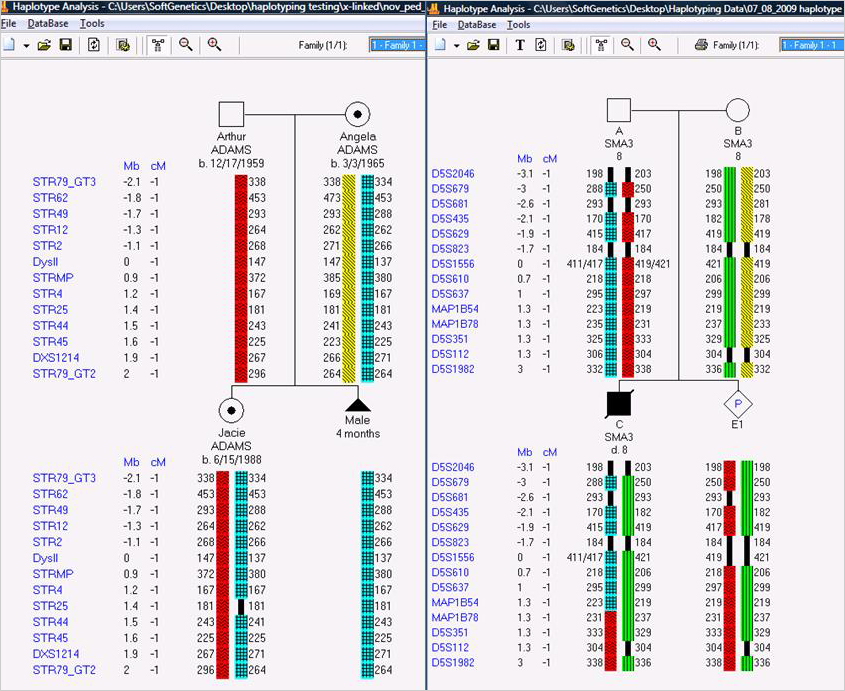
Figure 1: Personal information Phase assignment Carrier status (left). Phase assignment Cross-over in child 1 (right). Standard symbols are used for male, female, deceased and pregnancy.
Application Notes:
MLPA® is the registered trademark of MRC Holland













