MLVA Analysis (Multiple-Locus Variable Number Tandem Repeat Analysis) of Microbial Populations with Applications in Food Safety, Source Tracking of Food Borne Illnesses and Disease Transmission Routes
MLVA (Multiple-Locus Variable number tandem repeat Analysis is a molecular typing method using VNTRs (Variable Number Tandem Repeat), similar to simple sequence repeat (SSR) and STR (Short Tandem Repeat, microsatellite) analysis in eukaryotes, and is used in research ranging from transmission route studies, antibiotic resistance (such as MRSA) to food safety (Salmonela testing) and microbial source tracking. MLVA has better resolution than other microbial identification methods (typing of capsular polysaccharides and surface protein, Spa-sequence typing), which enables it to provide results that are easier to use in clustering analyses. MLVA is also less expensive and labor-intensive than multi-locus sequence typing [MLST] or pulsed-field gel electrophoresis. GeneMarker software provides fast, accurate genotyping of MLVA data from all capillary electrophoresis instruments – including ABI®PRISM, Applied Biosystems SeqStudio™, and Promega Spectrum Compact CE genetic analyzers and custom primers or commercially available chemistries– and is recommended for MLVA by European Staphylococcal reference laboratories. The flexible panel editor in GeneMarker software provides easy customization of panels for commercial or custom chemistries (primer sets).
Main Analysis Screen
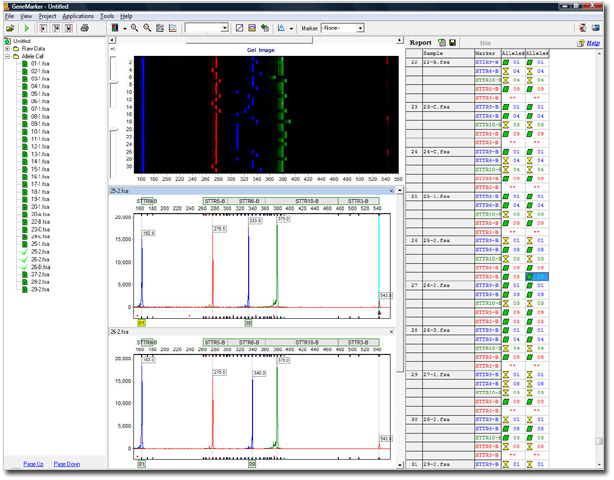
Figure 1: The screenshot above shows Linked Navigation between the electropherogram and allele table provides rapid review of results. Export report tables as .txt, .csv, .xlsx or image allele reports of the electropherograms with peak tables (.pdf, .png., jpeg).
MLPA® is the registered trademark of MRC Holland













