Rapid Concordance Testing and Determining a Consensus Genotype by Comparing Replicate Samples
Preparing sample replicates is a common practice in clinical research, wild and domesticated animal genetic profiling, and ecology/environmental research. Typically samples are amplified using a commercially available PCR kit or custom chemistry and the products then undergo capillary electrophoresis on an ABI®PRISM, Applied Biosystems SeqStudio™, and Promega Spectrum Compact CE genetic analyzer This technique lessens the impact of allele dropout and other common artifacts, and allows the analyst to confidently deduce a consensus genotype.
The replicate comparison tool groups and compares replicates of the same sample. The results are displayed in the report table, and the alignment of the replicates is displayed in the electropherogram (Figure 1). The comparison settings are flexible: replicates can be compared simply by the allele call at a given marker, or by additional parameters, such as peak size, height, and/or area. GeneMarker software supports several digital and hard copy reporting formats, including text, excel, and customizable print reports.
Figures 1 – 5 (below) provide an overview of the Replicate Comparison Tool in GeneMarker software.
Replicate Comparison View
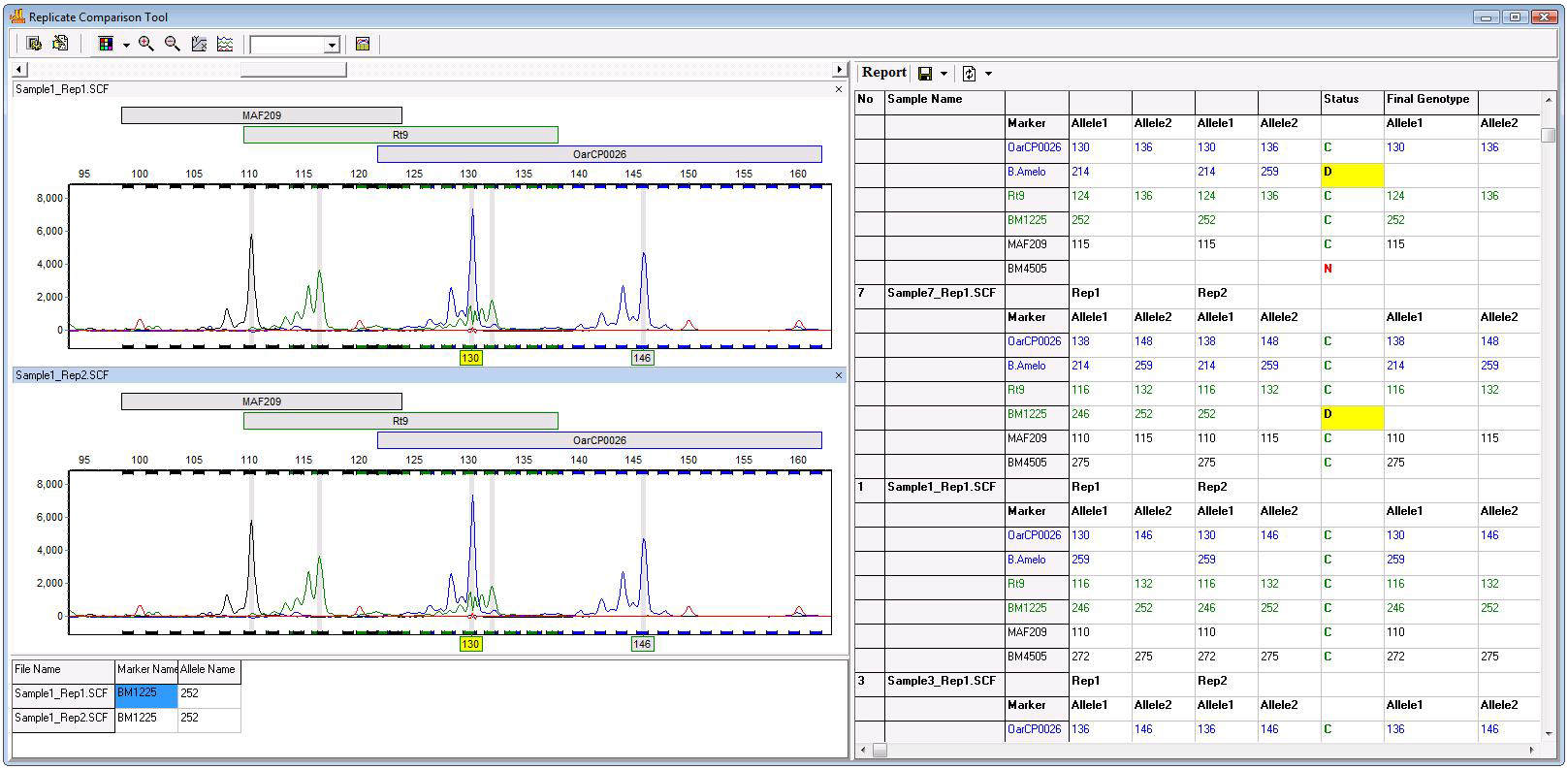
Figure 1: Two or more replicates are compared per the parameters set by the analyst (figure 2). Electrophoergrams are displayed at the left and a report table indicating any non-concordant alleles is displayed at the right.
Select the appropriate parameters
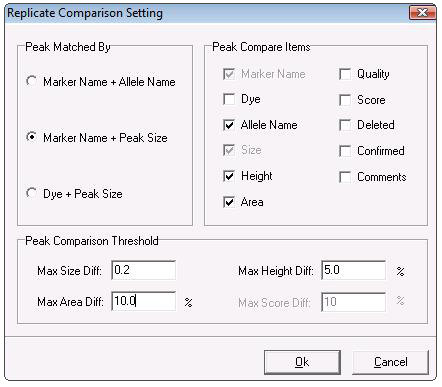
Figure 2: Use the Replicate Comparison Settings window to choose which parameters to compare for each replicate. For some parameters (size, area, height, and score) the user may set a range of allowable values.
A closer look at the Comparison Table

Figure 3: The report table displays the allele call for each replicate at a given marker, the results of the comparison of those calls (concordant, discordant, or null), and the consensus genotype at the specified marker. This table can be exported as a tab-delimited text file. The informative report table displays the allele calls for each replicate at the specified marker, the “status” of that marker, (described below), and the Final Genotype at that locus. The status column for a specified marker will display a C for Concordant if the replicates have the same call, a D for Discordant, if the call is different, and N, for Null, if only one replicate has allele calls (i.e. the other replicates failed). The report table is linked with the electropherogram; simply click on an allele or the corresponding status to be taken to that locus in the electropherogram. For markers with C and N calls, the final genotype column is automatically filled with the available genotype. In cases where the status is D, the user may manually select the genotype of that locus from a dropdown menu. The final report may then be exported as a tab-delimited text file.
Resolve Discordant Allele Calls
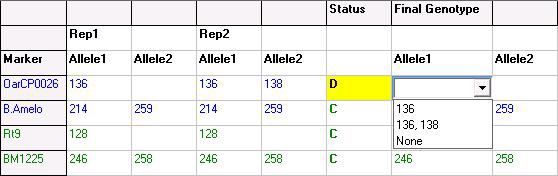
Figure 4: For discordant (conflicting) allele calls, the user may review the electropherograms displayed at the left side of the table and manually select the genotype of the specified locus.
The replicate comparison can accommodate replicates of 2 – 8 files
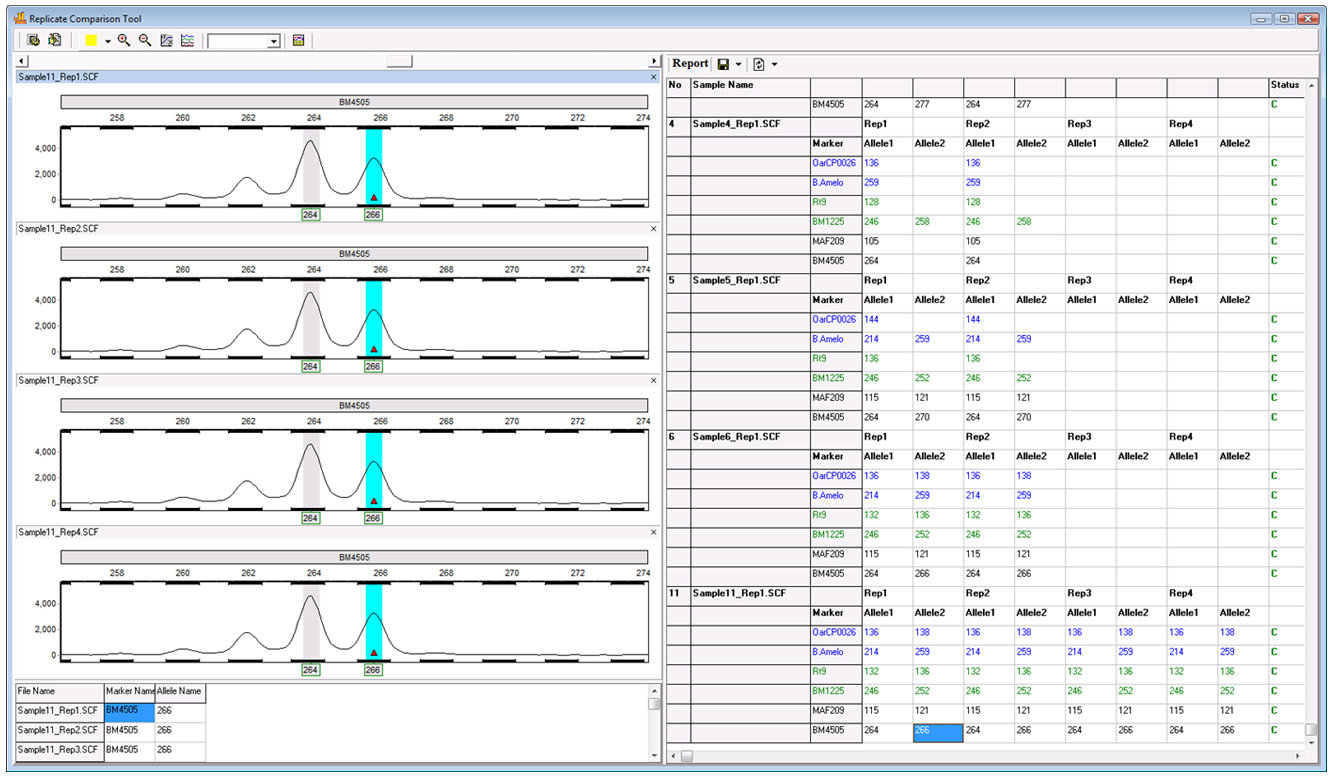
Figure 5: The replicate comparison tool can comfortably accommodate more than two replicates in each group. Shown above is a project in which four replicates were processed for each sample.
Application Notes:
MLPA® is the registered trademark of MRC Holland













