SNaPshot® SBE Analysis (Single Base Extension) Data Analysis to Detect Mutations in Clinical and Anthropology Research
SNaPshot chemistries are used to detect Single Nucleotide Polymorphisms (SNPs) in clinical research (such as with KRAS and BRAF mutation analysis and HIV drug resistance) or in ancestry prediction research. An unlabeled primer with its 3' end directly flanking the SNP is extended one nucleotide by Taq polymerase, and fluorescently-labeled ddNTPs complementary to the polymorphic base are added. The resulting fragment is one nucleotide longer, but the observed fragment size from electrophoresis will be greater than expected due to the influence of the fluorescent dye on the electrophoretic mobility of these small fragments. SNPs can be identified by the one- or two-color peaks associated with the incorporated labeled ddNTP and the length of the primer.
GeneMarker® software is an excellent tool to determine SNP genotypes from SBE SNaPshot data from capillary electrophoresis systems, including ABI®PRISM, Applied Biosystems® SeqStudio,, and Promega Spectrum Compact CE Systems genetic analyzers.
Our user-friendly GeneMarker software displays two-color SNPs on one trace, which is essential for analysis of SNaPshot CE data. GeneMarker software contains robust size calling algorithms that resolve fragment lengths to less than one base pair with high efficiency allele calling, and adjusts for mobility shifts of small fragments.
SNaPshot Analysis
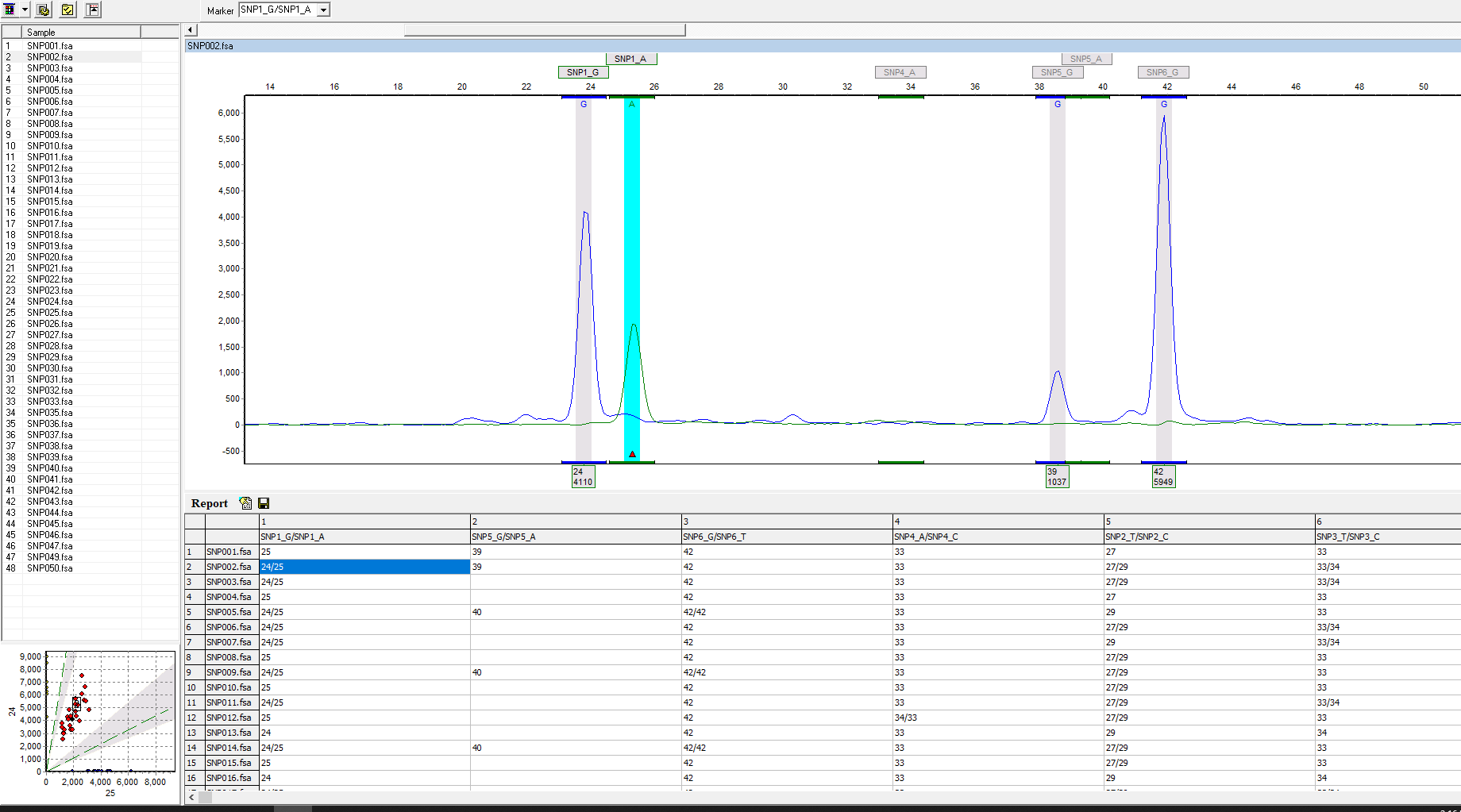
Figure 1: The analysis screen displays two-color SNPs on one trace and a SNP table. Linked navigation highlights the corresponding cell when the peak is selected in the electropherogram, as seen in this figure with a G to A mutation.
Application Notes:
MLPA® is the registered trademark of MRC Holland













