Kinship Analysis of Wild Populations and data base searching
GeneMarker® software is an efficient, user-friendly alternative to GeneMapper® software for wildlife and plant genotyping. The embedded Kinship Analysis and database search module which enables ecologists and other wildlife researchers to easily identify kinships in natural plant and animal populations, poaching cases, livestock breeding programs, and aquaculture without transferring allele calls to a separate spread sheet or database. The Kinship Analysis tool provides a report table with probabilities and likelihood ratios across three generations for sample pairs. The rigorous statistical analysis (Brenner, 1997; Li and Sachs 1954) to determine levels of kinship uses identity by descent (IBD) equations. The GeneMarker software database search tool identifies samples with the same STR profile, calculates the random match probability, then identifies and ranks files with the highest likelihood ratio for each relationship level to the experimental sample. Genetic Analysis Parameters allow setting for mistyping or mutation. The Database accepts allele frequency tables (species-specific markers), genotypes from current projects or combined multiplex results, and previously archived genotype .txt or .cmf files, providing easy database updates.
Database Search Report
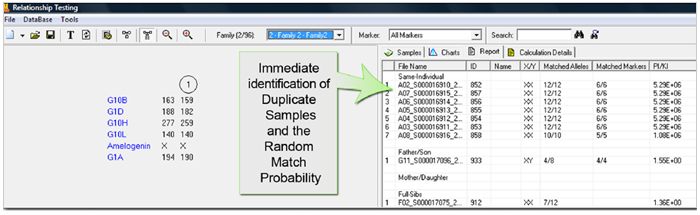
Figure 1: Users can do a Customize Search for animal or plant samples. Each individual file is represented as a node (circle – female, square = male). The genotype is listed below the node.
Kinship Analysis
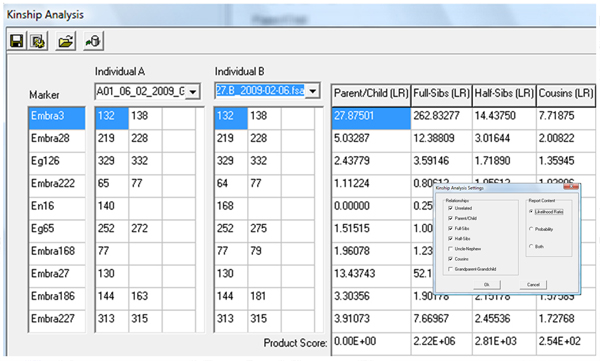
Figure 2: Kinship analysis settings enable users to customize the report format.
Application Notes:
- Kinship and Database Search: Animals
- Kinship and Database Search: Monoecious Plants and Invertebrates
- Plant & Animal Genome Kinship Poster
Webinars:
MLPA® is the registered trademark of MRC Holland













